Identification of structurally diverse menaquinone-binding antibiotics with in vivo activity against multidrug-resistant pathogens
- PMID: 34949828
- PMCID: PMC8732328
- DOI: 10.1038/s41564-021-01013-8
Identification of structurally diverse menaquinone-binding antibiotics with in vivo activity against multidrug-resistant pathogens
Abstract
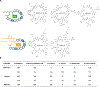
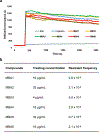
The emergence of multidrug-resistant bacteria poses a threat to global health and necessitates the development of additional in vivo active antibiotics with diverse modes of action. Directly targeting menaquinone (MK), which plays an important role in bacterial electron transport, is an appealing, yet underexplored, mode of action due to a dearth of MK-binding molecules. Here we combine sequence-based metagenomic mining with a motif search of bioinformatically predicted natural product structures to identify six biosynthetic gene clusters that we predicted encode MK-binding antibiotics (MBAs). Their predicted products (MBA1-6) were rapidly accessed using a synthetic bioinformatic natural product approach, which relies on bioinformatic structure prediction followed by chemical synthesis. Among these six structurally diverse MBAs, four make up two new MBA structural families. The most potent member of each new family (MBA3, MBA6) proved effective at treating methicillin-resistant Staphylococcus aureus infection in a murine peritonitis-sepsis model. The only conserved feature present in all MBAs is the sequence 'GXLXXXW', which we propose represents a minimum MK-binding motif. Notably, we found that a subset of MBAs were active against Mycobacterium tuberculosis both in vitro and in macrophages. Our findings suggest that naturally occurring MBAs are a structurally diverse and untapped class of mechanistically interesting, in vivo active antibiotics.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests
The authors declare no competing financial interests.
Figures








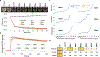
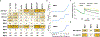


References
-
- Wellcome Trust and UK Government. Review on antimicrobial resistance – tackling drug-resistant infections globally: Final report and recommendations Wellcome Trust and UK Government; (2016).
-
- Brown ED & Wright GD Antibacterial drug discovery in the resistance era. Nature 529, 336–343 (2016). - PubMed
-
- Niu GQ & Li WL Next-generation drug discovery to combat antimicrobial resistance. Trends Biochem. Sci 44, 961–972 (2019). - PubMed
-
- Lewis K The science of antibiotic discovery. Cell 181, 29–45 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

