Aging disrupts circadian gene regulation and function in macrophages
- PMID: 34949832
- PMCID: PMC9704320
- DOI: 10.1038/s41590-021-01083-0
Aging disrupts circadian gene regulation and function in macrophages
Abstract
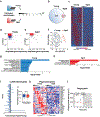
Aging is characterized by an increased vulnerability to infection and the development of inflammatory diseases, such as atherosclerosis, frailty, cancer and neurodegeneration. Here, we find that aging is associated with the loss of diurnally rhythmic innate immune responses, including monocyte trafficking from bone marrow to blood, response to lipopolysaccharide and phagocytosis. This decline in homeostatic immune responses was associated with a striking disappearance of circadian gene transcription in aged compared to young tissue macrophages. Chromatin accessibility was significantly greater in young macrophages than in aged macrophages; however, this difference did not explain the loss of rhythmic gene transcription in aged macrophages. Rather, diurnal expression of Kruppel-like factor 4 (Klf4), a transcription factor (TF) well established in regulating cell differentiation and reprogramming, was selectively diminished in aged macrophages. Ablation of Klf4 expression abolished diurnal rhythms in phagocytic activity, recapitulating the effect of aging on macrophage phagocytosis. Examination of individuals harboring genetic variants of KLF4 revealed an association with age-dependent susceptibility to death caused by bacterial infection. Our results indicate that loss of rhythmic Klf4 expression in aged macrophages is associated with disruption of circadian innate immune homeostasis, a mechanism that may underlie age-associated loss of protective immune responses.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures



Comment in
-
A broken immune clock in old macrophages.Nat Rev Immunol. 2022 Feb;22(2):74-75. doi: 10.1038/s41577-021-00674-0. Nat Rev Immunol. 2022. PMID: 34983967 No abstract available.
-
Aging alters rhythms in immunity.Nat Immunol. 2022 Feb;23(2):153-154. doi: 10.1038/s41590-021-01099-6. Nat Immunol. 2022. PMID: 35079158 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

