Characterization of the liver immune microenvironment in liver biopsies from patients with chronic HBV infection
- PMID: 34950863
- PMCID: PMC8671126
- DOI: 10.1016/j.jhepr.2021.100388
Characterization of the liver immune microenvironment in liver biopsies from patients with chronic HBV infection
Abstract
Background & aims: We aim to describe the liver immune microenvironment by analyzing liver biopsies from patients with chronic HBV infection (CHB). Host immune cell signatures and their corresponding localization were characterized by analyzing the intrahepatic transcriptome in combination with a custom multiplex immunofluorescence panel.
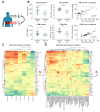
Method: Matching FFPE and fresh frozen liver biopsies were collected from immune active patients within the open-label phase IV study GS-US-174-0149. RNA-Seq was conducted on 53 CHB liver biopsies from 46 patients. Twenty-eight of the 53 samples had matched FFPE biopsies and were stained with a 12-plex panel including cell segmentation, immune and viral biomarkers. Corresponding serum samples were screened using the MSD Human V-plex Screen Service to identify peripheral correlates for the immune microenvironment.
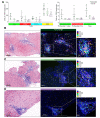
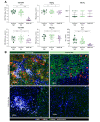
Results: Using unsupervised clustering of the transcriptome, we reveal two unique liver immune signatures classified as immune high and immune low based on the quantification of the liver infiltrate gene signatures. Multiplex immunofluorescence analysis demonstrated large periportal lymphoid aggregates in immune high samples consisting of CD4 and CD8 T cells, B cells and macrophages. Differentiation of the high and low immune microenvironments was independent of HBeAg status and peripheral viral antigen levels. In addition, longitudinal analysis indicates that treatment and normalization of ALT correlates with a decrease in liver immune infiltrate and inflammation. Finally, we screened a panel of peripheral biomarkers and identified ICAM-1 and CXCL10 as biomarkers that strongly correlate with these unique immune microenvironments.
Conclusion: These data provide a description of immune phenotypes in patients with CHB and show that immune responses are downregulated in the liver following nucleotide analogue treatment. This may have important implications for both the safety and efficacy of immune modulator programs aimed at HBV cure.
Lay summary: Liver biopsies from patients with chronic hepatitis B were submitted to RNA-Seq and multiplex immunofluorescence and identified two different liver immune microenvironments: immune high and immune low. Immune high patients showed elevated immune pathways, including interferon signaling pathways, and increase presence of immune cells. Longitudinal analysis of biopsies from treatment experienced patients showed that treatment correlates with a marked decrease in inflammation and these findings may have important implications for both safety and efficacy of immune modulator programs for HBV cure.
Keywords: ALT, alanine aminotransferase; BCR, B-cell receptor; CHB, chronic HBV infection; Chronic HBV; DEG, differentially expressed gene; FFPE, formalin-fixed paraffin-embedded; Hepatitis B; IHC, immunohistochemistry; Immune Microenvironment; Intrahepatic transcriptome; PEG-IFNα, pegylated-interferon-α; TCR, T-cell receptor; TDF, tenofovir disoproxil fumarate; TLS, tertiary lymphoid structures; mIF, multiplex immunofluorescence; multiplex immunofluorescence; ssGSEA, single sample gene set enrichment analysis.
© 2021 The Author(s).
Conflict of interest statement
All authors are either employees of Gilead Sciences or funded by Gilead Sciences for clinical trial sample collection. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures





References
-
- Organization WH . 2017. Hepatitis B.https://www.who.int/news-room/fact-sheets/detail/hepatitis-b [cited; Available from:
-
- Iloeje U.H., Yang H.I., Jen C.L., Su J., Wang L.Y., You S.L., et al. Risk and predictors of mortality associated with chronic hepatitis B infection. Clin Gastroenterol Hepatol. 2007;5:921–931. - PubMed
-
- Fattovich G., Stroffolini T., Zagni I., Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–S50. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

