A protocol for imaging microvilli biogenesis on the surface of cultured porcine kidney epithelial cell monolayers
- PMID: 34950883
- PMCID: PMC8672049
- DOI: 10.1016/j.xpro.2021.100998
A protocol for imaging microvilli biogenesis on the surface of cultured porcine kidney epithelial cell monolayers
Abstract
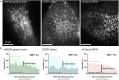
A key facet of epithelial differentiation is the assembly of actin-based protrusions known as microvilli, which amplify apical membrane surface area for various cell functions. To probe mechanisms of microvillus assembly, we developed a protocol using spinning disk confocal microscopy to directly visualize microvillus biogenesis on the surface of cultured porcine kidney epithelial cell monolayers engineered to express fluorescent proteins. This protocol offers access to the molecular details of individual protrusion growth events at high spatiotemporal resolution. For complete details on the use and execution of this protocol, please refer to Gaeta et al. (2021).
Keywords: Cell Biology; Cell culture; Cell-based Assays; Microscopy.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Baas A.F., Kuipers J., van der Wel N.N., Batlle E., Koerten H.K., Peters P.J., Clevers H.C. Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell. 2004;116:457–466. - PubMed
-
- Melak M., Plessner M., Grosse R. Actin visualization at a glance. J. Cell Sci. 2017;130:525–530. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

