New interpretable machine-learning method for single-cell data reveals correlates of clinical response to cancer immunotherapy
- PMID: 34950900
- PMCID: PMC8672150
- DOI: 10.1016/j.patter.2021.100372
New interpretable machine-learning method for single-cell data reveals correlates of clinical response to cancer immunotherapy
Abstract
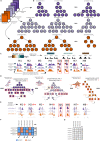
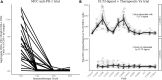
We introduce a new method for single-cell cytometry studies, FAUST, which performs unbiased cell population discovery and annotation. FAUST processes experimental data on a per-sample basis and returns biologically interpretable cell phenotypes, making it well suited for the analysis of complex datasets. We provide simulation studies that compare FAUST with existing methodology, exemplifying its strength. We apply FAUST to data from a Merkel cell carcinoma anti-PD-1 trial and discover pre-treatment effector memory T cell correlates of outcome co-expressing PD-1, HLA-DR, and CD28. Using FAUST, we then validate these correlates in cryopreserved peripheral blood mononuclear cell samples from the same study, as well as an independent CyTOF dataset from a published metastatic melanoma trial. Finally, we show how FAUST's phenotypes can be used to perform cross-study data integration in the presence of diverse staining panels. Together, these results establish FAUST as a powerful new approach for unbiased discovery in single-cell cytometry.
Keywords: algorithms; bioinformatics; cancer; immunology; single-cell; statistics & probability.
© 2021 The Authors.
Conflict of interest statement
A patent for the application of the FAUST algorithm to cytometry datasets has been applied for on behalf of the Fred Hutchinson Cancer Research Center. The research described in this paper was completed while E.G. was conducting research and working at the Fred Hutchinson Cancer Research Center. E.G. declares ownership interest in Ozette Technologies and was an employee of Ozette Technologies when the manuscript was revised to respond to peer review. G.F. has received consulting income from Takeda and research support from Janssen Pharmaceuticals and declares ownership interest in Ozette Technologies. R.G. has received consulting income from Juno Therapeutics, Takeda, Infotech Soft, and Celgene, has received research support from Janssen Pharmaceuticals and Juno Therapeutics, and declares ownership in Ozette Technologies and Modulus Therapeutics. Trial funds for CITN-07 were in part provided by Celldex. Trial funds for CITN-09 were in part provided by Merck.
Figures




References
-
- Grégori G., Patsekin V., Rajwa B., Jones J., Ragheb K., Holdman C., Robinson J.P. Hyperspectral cytometry at the single-cell level using a 32-channel photodetector. Cytometry Part A. 2012;81:35–44. - PubMed
-
- Saeys Y., Van Gassen S., Lambrecht B.N. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat. Rev. Immunol. 2016;16:449. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

