Molecular determinants of phase separation for Drosophila DNA replication licensing factors
- PMID: 34951585
- PMCID: PMC8813052
- DOI: 10.7554/eLife.70535
Molecular determinants of phase separation for Drosophila DNA replication licensing factors
Abstract
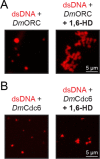
Liquid-liquid phase separation (LLPS) of intrinsically disordered regions (IDRs) in proteins can drive the formation of membraneless compartments in cells. Phase-separated structures enrich for specific partner proteins and exclude others. Previously, we showed that the IDRs of metazoan DNA replication initiators drive DNA-dependent phase separation in vitro and chromosome binding in vivo, and that initiator condensates selectively recruit replication-specific partner proteins (Parker et al., 2019). How initiator IDRs facilitate LLPS and maintain compositional specificity is unknown. Here, using Drosophila melanogaster (Dm) Cdt1 as a model initiation factor, we show that phase separation results from a synergy between electrostatic DNA-bridging interactions and hydrophobic inter-IDR contacts. Both sets of interactions depend on sequence composition (but not sequence order), are resistant to 1,6-hexanediol, and do not depend on aromaticity. These findings demonstrate that distinct sets of interactions drive condensate formation and specificity across different phase-separating systems and advance efforts to predict IDR LLPS propensity and partner selection a priori.
Keywords: D. melanogaster; DNA binding proteins; DNA replication; biochemistry; chemical biology; intrinsically disordered proteins; liquid phase condensates.
© 2021, Parker et al.
Conflict of interest statement
MP, JK, AH No competing interests declared, JB, MB Reviewing editor, eLife
Figures















References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

