Text Mining of Adverse Events in Clinical Trials: Deep Learning Approach
- PMID: 34951601
- PMCID: PMC8742206
- DOI: 10.2196/28632
Text Mining of Adverse Events in Clinical Trials: Deep Learning Approach
Abstract
Background: Pharmacovigilance and safety reporting, which involve processes for monitoring the use of medicines in clinical trials, play a critical role in the identification of previously unrecognized adverse events or changes in the patterns of adverse events.
Objective: This study aims to demonstrate the feasibility of automating the coding of adverse events described in the narrative section of the serious adverse event report forms to enable statistical analysis of the aforementioned patterns.
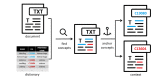
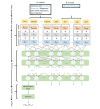
Methods: We used the Unified Medical Language System (UMLS) as the coding scheme, which integrates 217 source vocabularies, thus enabling coding against other relevant terminologies such as the International Classification of Diseases-10th Revision, Medical Dictionary for Regulatory Activities, and Systematized Nomenclature of Medicine). We used MetaMap, a highly configurable dictionary lookup software, to identify the mentions of the UMLS concepts. We trained a binary classifier using Bidirectional Encoder Representations from Transformers (BERT), a transformer-based language model that captures contextual relationships, to differentiate between mentions of the UMLS concepts that represented adverse events and those that did not.
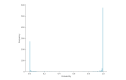
Results: The model achieved a high F1 score of 0.8080, despite the class imbalance. This is 10.15 percent points lower than human-like performance but also 17.45 percent points higher than that of the baseline approach.
Conclusions: These results confirmed that automated coding of adverse events described in the narrative section of serious adverse event reports is feasible. Once coded, adverse events can be statistically analyzed so that any correlations with the trialed medicines can be estimated in a timely fashion.
Keywords: classification; deep learning; machine learning; natural language processing.
©Daphne Chopard, Matthias S Treder, Padraig Corcoran, Nagheen Ahmed, Claire Johnson, Monica Busse, Irena Spasic. Originally published in JMIR Medical Informatics (https://medinform.jmir.org), 24.12.2021.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures






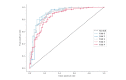
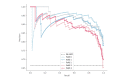
References
-
- Data Mining at FDA - White Paper. US Food and Drug Administration. 2018. [2021-12-11]. https://www.fda.gov/science-research/data-mining/data-mining-fda-white-p... .
-
- Botsis T, Nguyen MD, Woo EJ, Markatou M, Ball R. Text mining for the Vaccine Adverse Event Reporting System: medical text classification using informative feature selection. J Am Med Inform Assoc. 2011;18(5):631–8. doi: 10.1136/amiajnl-2010-000022. http://europepmc.org/abstract/MED/21709163 amiajnl-2010-000022 - DOI - PMC - PubMed
-
- Chee BW, Berlin R, Schatz B. Predicting adverse drug events from personal health messages. AMIA Annu Symp Proc. 2011;2011:217–26. http://europepmc.org/abstract/MED/22195073 - PMC - PubMed
-
- Botsis T, Buttolph T, Nguyen MD, Winiecki S, Woo EJ, Ball R. Vaccine adverse event text mining system for extracting features from vaccine safety reports. J Am Med Inform Assoc. 2012;19(6):1011–8. doi: 10.1136/amiajnl-2012-000881. http://europepmc.org/abstract/MED/22922172 amiajnl-2012-000881 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

