ATRX loss promotes immunosuppressive mechanisms in IDH1 mutant glioma
- PMID: 34951647
- PMCID: PMC9159463
- DOI: 10.1093/neuonc/noab292
ATRX loss promotes immunosuppressive mechanisms in IDH1 mutant glioma
Abstract
Background: ATRX inactivation occurs with IDH1R132H and p53 mutations in over 80% of Grades II/III astrocytomas. It is believed that ATRX loss contributes to oncogenesis by dysregulating epigenetic and telomere mechanisms but effects on anti-glioma immunity have not been explored. This paper examines how ATRX loss contributes to the malignant and immunosuppressive phenotypes of IDH1R132H/p53mut glioma cells and xenografts.
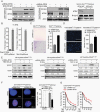
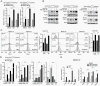
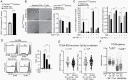
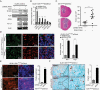
Methods: Isogenic astrocytoma cells (+/-IDH1R132H/+/-ATRXloss) were established in p53mut astrocytoma cell lines using lentivirus encoding doxycycline-inducible IDH1R132H, ATRX shRNA, or Lenti-CRISPR/Cas9 ATRX. Effects of IDH1R132H+/-ATRXloss on cell migration, growth, DNA repair, and tumorigenicity were evaluated by clonal growth, transwell and scratch assays, MTT, immunofluorence and immunoblotting assays, and xenograft growth. Effects on the expression and function of modulators of the immune microenvironment were quantified by qRT-PCR, immunoblot, T-cell function, macrophage polarization, and flow cytometry assays. Pharmacologic inhibitors were used to examine epigenetic drivers of the immunosuppressive transcriptome of IDH1R132H/p53mut/ATRXloss cells.
Results: Adding ATRX loss to the IDH1R132H/p53mut background promoted astrocytoma cell aggressiveness, induced expression of BET proteins BRD3/4 and an immune-suppressive transcriptome consisting of up-regulated immune checkpoints (e.g., PD-L1, PD-L2) and altered cytokine/chemokine profiles (e.g., IL33, CXCL8, CSF2, IL6, CXCL9). ATRX loss enhanced the capacity of IDH1R132H/p53mut cells to induce T-cell apoptosis, tumorigenic/anti-inflammatory macrophage polarization and Treg infiltration. The transcriptional and biological immune-suppressive responses to ATRX loss were enhanced by temozolomide and radiation and abrogated by pharmacologic BET inhibition.
Conclusions: ATRX loss activates a BRD-dependent immune-suppressive transcriptome and immune escape mechanism in IDH1R132H/p53mut astrocytoma cells.
Keywords: ATRX loss; BRD-dependent immune-suppressive transcriptome; IDH1 mutant astrocytoma; glioma immune microenvironment.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


Comment in
-
Loss of ATRX suppresses anti-tumor immunity.Neuro Oncol. 2022 Jun 1;24(6):901-902. doi: 10.1093/neuonc/noac059. Neuro Oncol. 2022. PMID: 35235678 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

