Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance
- PMID: 34951957
- PMCID: PMC8847337
- DOI: 10.1016/j.ccell.2021.12.001
Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance
Abstract
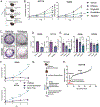
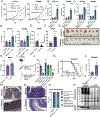
Microbial dysbiosis is a colorectal cancer (CRC) hallmark and contributes to inflammation, tumor growth, and therapy response. Gut microbes signal via metabolites, but how the metabolites impact CRC is largely unknown. We interrogated fecal metabolites associated with mouse models of colon tumorigenesis with varying mutational load. We find that microbial metabolites from healthy mice or humans are growth-repressive, and this response is attenuated in mice and patients with CRC. Microbial profiling reveals that Lactobacillus reuteri and its metabolite, reuterin, are downregulated in mouse and human CRC. Reuterin alters redox balance, and reduces proliferation and survival in colon cancer cells. Reuterin induces selective protein oxidation and inhibits ribosomal biogenesis and protein translation. Exogenous Lactobacillus reuteri restricts colon tumor growth, increases tumor reactive oxygen species, and decreases protein translation in vivo. Our findings indicate that a healthy microbiome and specifically, Lactobacillus reuteri, is protective against CRC through microbial metabolite exchange.
Keywords: Lactobacillus reuteri; Microbiome; Reuterin; colorectal cancer; metabolites; protein oxidation.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Comment in
-
Lactobacillus reuteri-an old acquaintance takes on a new task in colorectal tumor surveillance.Cancer Cell. 2022 Feb 14;40(2):125-127. doi: 10.1016/j.ccell.2022.01.014. Cancer Cell. 2022. PMID: 35167822
References
-
- Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, and Bray F (2017). Global patterns and trends in colorectal cancer incidence and mortality. Gut 66, 683. - PubMed
-
- Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, Moryama EH, Copeland JK, Surendra A, Kumar S, et al. (2014). Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 158(2):288–299. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA148828/CA/NCI NIH HHS/United States
- R01 DK095201/DK/NIDDK NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- R01 CA245546/CA/NCI NIH HHS/United States
- R01 CA248160/CA/NCI NIH HHS/United States
- T32 GM007863/GM/NIGMS NIH HHS/United States
- I01 BX004842/BX/BLRD VA/United States
- R37 CA237421/CA/NCI NIH HHS/United States
- R01 NS099280/NS/NINDS NIH HHS/United States
- F30 CA257292/CA/NCI NIH HHS/United States
- P50 HD104463/HD/NICHD NIH HHS/United States
- P30 DK089503/DK/NIDDK NIH HHS/United States
- P50 CA130810/CA/NCI NIH HHS/United States
- R01 CA244931/CA/NCI NIH HHS/United States
- P30 DK034933/DK/NIDDK NIH HHS/United States
- F31 NS127371/NS/NINDS NIH HHS/United States
- F99 CA264414/CA/NCI NIH HHS/United States
- F31 CA247457/CA/NCI NIH HHS/United States
- U24 DK097153/DK/NIDDK NIH HHS/United States
- T32 GM007315/GM/NIGMS NIH HHS/United States
- R01 CA215607/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

