Genomic analysis of SARS-CoV-2 variants of concern identified from the ChAdOx1 nCoV-19 immunized patients from Southwest part of Bangladesh
- PMID: 34952247
- PMCID: PMC8688836
- DOI: 10.1016/j.jiph.2021.12.002
Genomic analysis of SARS-CoV-2 variants of concern identified from the ChAdOx1 nCoV-19 immunized patients from Southwest part of Bangladesh
Abstract
Background: Bangladesh introduced ChAdOx1 nCoV-19 since February, 2021 and in six months, only a small population (12.8%) received either one or two dose of vaccination like other low-income countries. The COVID-19 infections were continued to roll all over the places although the information on genomic variations of SARS-CoV-2 between both immunized and unimmunized group was unavailable. The objective of this study was to compare the proportion of immune escaping variants between those groups.
Methods: A total of 4718 nasopharygeal samples were collected from March 1 until April 15, 2021, of which, 834 (18%) were SARS-CoV-2 positive. The minimum sample size was calculated as 108 who were randomly selected for telephone interview and provided consent. The prevalence of SARS-CoV-2 variants and disease severity among both immunized and unimmunized groups was measured. A total of 63 spike protein sequences and 14 whole-genome sequences were performed from both groups and phylogenetic reconstruction and mutation analysis were compared.
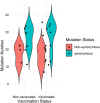
Results: A total of 40 respondents (37%, N = 108) received single-dose and 2 (2%) received both doses of ChAdOx1 nCoV-19 vaccine, which significantly reduce dry cough, loss of appetite and difficulties in breathing compared to none. There was no significant difference in hospitalization, duration of hospitalization or reduction of other symptoms like running nose, muscle pain, shortness of breathing or generalized weakness between immunized and unimmunized groups. Spike protein sequence assumed 21 (87.5%) B.1.351, one B.1.526 and two 20B variants in immunized group compared to 27 (69%) B.1.351, 5 (13%) B.1.1.7, 4 (10%) 20B, 2 B.1.526 and one B.1.427 variant in unimmunized group. Whole genome sequence analysis of 14 cases identified seven B.1.351 Beta V2, three B.1.1.7 Alpha V1, one B.1.526 Eta and the rest three 20B variants.
Conclusion: Our study observed that ChAdOx1 could not prevent the new infection or severe COVID-19 disease outcome with single dose while the infections were mostly caused by B.1.351 variants in Bangladesh.
Keywords: B.1.351; COVID-19; ChAdOx1 nCoV-19; Oxford-AstraZeneca; South African variant.
Copyright © 2021 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
Similar articles
-
Genomic characterization of the dominating Beta, V2 variant carrying vaccinated (Oxford-AstraZeneca) and nonvaccinated COVID-19 patient samples in Bangladesh: A metagenomics and whole-genome approach.J Med Virol. 2022 Apr;94(4):1670-1688. doi: 10.1002/jmv.27537. Epub 2022 Jan 7. J Med Virol. 2022. PMID: 34939673
-
Heterologous ChAdOx1 nCoV-19 and BNT162b2 prime-boost vaccination elicits potent neutralizing antibody responses and T cell reactivity against prevalent SARS-CoV-2 variants.EBioMedicine. 2022 Jan;75:103761. doi: 10.1016/j.ebiom.2021.103761. Epub 2021 Dec 17. EBioMedicine. 2022. PMID: 34929493 Free PMC article. Clinical Trial.
-
Antibody responses after a single dose of ChAdOx1 nCoV-19 vaccine in healthcare workers previously infected with SARS-CoV-2.EBioMedicine. 2021 Aug;70:103523. doi: 10.1016/j.ebiom.2021.103523. Epub 2021 Aug 12. EBioMedicine. 2021. PMID: 34391088 Free PMC article.
-
Herpes simplex encephalitis following ChAdOx1 nCoV-19 vaccination: a case report and review of the literature.BMC Infect Dis. 2022 Mar 3;22(1):217. doi: 10.1186/s12879-022-07186-9. BMC Infect Dis. 2022. PMID: 35241013 Free PMC article. Review.
-
SARS-CoV-2 Variants and COVID-19 in Bangladesh-Lessons Learned.Viruses. 2024 Jul 4;16(7):1077. doi: 10.3390/v16071077. Viruses. 2024. PMID: 39066238 Free PMC article. Review.
Cited by
-
Alternative genome sequencing approaches of SARS-CoV-2 using Ion AmpliSeq Technology.MethodsX. 2024 Mar 11;12:102646. doi: 10.1016/j.mex.2024.102646. eCollection 2024 Jun. MethodsX. 2024. PMID: 38524302 Free PMC article.
-
Microbiome analysis revealing microbial interactions and secondary bacterial infections in COVID-19 patients comorbidly affected by Type 2 diabetes.J Med Virol. 2023 Jan;95(1):e28234. doi: 10.1002/jmv.28234. Epub 2022 Nov 1. J Med Virol. 2023. PMID: 36258280 Free PMC article.
-
Temporal dynamics and fatality of SARS-CoV-2 variants in Bangladesh.Health Sci Rep. 2023 Apr 17;6(4):e1209. doi: 10.1002/hsr2.1209. eCollection 2023 Apr. Health Sci Rep. 2023. PMID: 37077184 Free PMC article.
-
Some common deleterious mutations are shared in SARS-CoV-2 genomes from deceased COVID-19 patients across continents.Sci Rep. 2023 Oct 30;13(1):18644. doi: 10.1038/s41598-023-45517-1. Sci Rep. 2023. PMID: 37903828 Free PMC article.
-
Emergence of SARS-CoV-2 Variants Are Induced by Coinfections With Dengue.Bioinform Biol Insights. 2024 Sep 11;18:11779322241272399. doi: 10.1177/11779322241272399. eCollection 2024. Bioinform Biol Insights. 2024. PMID: 39290577 Free PMC article.
References
-
- Corona.gov.bd. corona.gov.bd press-releases December 2020. https://corona.gov.bd/storage/press-releases/December2020/KsvQiLw3eIdbv2.... 2020.
-
- DGHS. Coronavirus COVID-19 Dashboard, http://dashboard.dghs.gov.bd/webportal/pages/covid19.php. 2021. [Accessed 18 November 2021].
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

