Carbon catabolite repression involves physical interaction of the transcription factor CRE1/CreA and the Tup1-Cyc8 complex in Penicillium oxalicum and Trichoderma reesei
- PMID: 34952627
- PMCID: PMC8710005
- DOI: 10.1186/s13068-021-02092-9
Carbon catabolite repression involves physical interaction of the transcription factor CRE1/CreA and the Tup1-Cyc8 complex in Penicillium oxalicum and Trichoderma reesei
Abstract
Background: Cellulolytic enzyme production in filamentous fungi requires a release from carbon catabolite repression (CCR). The protein CRE1/CreA (CRE = catabolite responsive element) is a key transcription factor (TF) that is involved in CCR and represses cellulolytic gene expression. CRE1/CreA represents the functional equivalent of Mig1p, an important Saccharomyces cerevisiae TF in CCR that exerts its repressive effect by recruiting a corepressor complex Tup1p-Cyc8p. Although it is known from S. cerevisiae that CRE1/CreA might repress gene expression via interacting with the corepressor complex Tup1-Cyc8, this mechanism is unconfirmed in other filamentous fungi, since the physical interaction has not yet been verified in these organisms. The precise mechanism on how CRE1/CreA achieves transcriptional repression after DNA binding remains unknown.
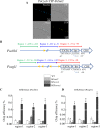
Results: The results from tandem affinity purification and bimolecular fluorescence complementation revealed a direct physical interaction between the TF CRE1/CreA and the complex Tup1-Cyc8 in the nucleus of cellulolytic fungus Trichoderma reesei and Penicillium oxalicum. Both fungi have the ability to secrete a complex arsenal of enzymes to synergistically degrade lignocellulosic materials. In P. oxalicum, the protein PoCyc8, a subunit of complex Tup1-Cyc8, interacts directly with TF PoCreA and histone H3 lysine 36 (H3K36) methyltransferase PoSet2 in the nucleus. The di-methylation level of H3K36 in the promoter of prominent cellulolytic genes (cellobiohydrolase-encoding gene Pocbh1/cel7A and endoglucanase-encoding gene Poegl1/cel7B) is positively correlated with the expression levels of TF PoCreA. Since the methylation of H3K36 was also demonstrated to be a repression marker of cellulolytic gene expression, it appears feasible that the cellulolytic genes are repressed via PoCreA-Tup1-Cyc8-Set2-mediated transcriptional repression.
Conclusion: This study verifies the long-standing conjecture that the TF CRE1/CreA represses gene expression by interacting with the corepressor complex Tup1-Cyc8 in filamentous fungi. A reasonable explanation is proposed that PoCreA represses gene expression by recruiting complex PoTup1-Cyc8. Histone methyltransferase Set2, which methylates H3K36, is also involved in the regulatory network by interacting with PoCyc8. The findings contribute to the understanding of CCR mechanism in filamentous fungi and could aid in biotechnologically relevant enzyme production.
Keywords: CCR; CRE1; Cellulases; Penicillium; Transcription factor; Trichoderma.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Isikgor FH, Becer CR. Lignocellulosic biomass: a sustainable platform for the production of bio-based chemicals and polymers. Polym Chem-Uk. 2015;6:4497–4559.
-
- Berner RA. The long-term carbon cycle, fossil fuels and atmospheric composition. Nature. 2003;426:323–326. - PubMed
-
- Lynd LR, Liang X, Biddy MJ, Allee A, Cai H, Foust T, et al. Cellulosic ethanol: status and innovation. Curr Opin Biotech. 2017;45:202–211. - PubMed
-
- Mäkelä MR, Donofrio N, de Vries RP. Plant biomass degradation by fungi. Fungal Genet Biol. 2014;72:2–9. - PubMed
-
- Antonieto ACC, Castro LD, Silva-Rocha R, Persinoti GF, Silva RN. Defining the genome-wide role of CRE1 during carbon catabolite repression in Trichoderma reesei using RNA-Seq analysis. Fungal Genet Biol. 2014;73:93–103. - PubMed
Grants and funding
- 2018YFA0900500/National Key Research and Development Project of China
- 32070077/National Natural Sciences Foundation of China
- 31370086/National Natural Sciences Foundation of China
- 31971387/National Natural Sciences Foundation of China
- ZR2019MC007/Shandong Provincial Natural Science Foundation, China
LinkOut - more resources
Full Text Sources
Miscellaneous

