Letter to the Editor: on the stability and internal consistency of component-wise sparse mixture regression-based clustering
- PMID: 34953466
- PMCID: PMC8769908
- DOI: 10.1093/bib/bbab532
Letter to the Editor: on the stability and internal consistency of component-wise sparse mixture regression-based clustering
Abstract
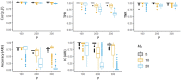
Understanding the relationship between molecular markers and a phenotype of interest is often obfuscated by patient-level heterogeneity. To address this challenge, Chang et al. recently published a novel method called Component-wise Sparse Mixture Regression (CSMR), a regression-based clustering method that promises to detect heterogeneous relationships between molecular markers and a phenotype of interest under high-dimensional settings. In this Letter to the Editor, we raise awareness to several issues concerning the assessment of CSMR in Chang et al., particularly its assessment in settings where the number of features, P, exceeds the study sample size, N, and advocate for additional metrics/approaches when assessing the performance of regression-based clustering methodologies.
Keywords: disease heterogeneity; mixture modeling; supervised learning.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

Comment on
-
Supervised clustering of high-dimensional data using regularized mixture modeling.Brief Bioinform. 2021 Jul 20;22(4):bbaa291. doi: 10.1093/bib/bbaa291. Brief Bioinform. 2021. PMID: 34293851 Free PMC article.
References
-
- Khalili A, Chen J. Variable selection in finite mixture of regression models. J Am Stat Assoc 2007;102(479):1025–38.
-
- Wang H, Leng C. Unified LASSO estimation by least squares approximation. J Am Stat Assoc 2007;102(479):1039–48.

