Isotopically nonstationary 13C metabolic flux analysis in resting and activated human platelets
- PMID: 34954086
- PMCID: PMC8905147
- DOI: 10.1016/j.ymben.2021.12.007
Isotopically nonstationary 13C metabolic flux analysis in resting and activated human platelets
Abstract
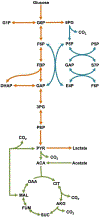
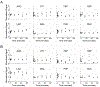
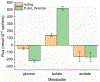
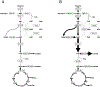
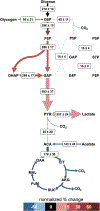
Platelet metabolism is linked to platelet hyper- and hypoactivity in numerous human diseases. Developing a detailed understanding of the link between metabolic shifts and platelet activation state is integral to improving human health. Here, we show the first application of isotopically nonstationary 13C metabolic flux analysis to quantitatively measure carbon fluxes in both resting and thrombin activated platelets. Metabolic flux analysis results show that resting platelets primarily metabolize glucose to lactate via glycolysis, while acetate is oxidized to fuel the tricarboxylic acid cycle. Upon activation with thrombin, a potent platelet agonist, platelets increase their uptake of glucose 3-fold. This results in an absolute increase in flux throughout central metabolism, but when compared to resting platelets they redistribute carbon dramatically. Activated platelets decrease relative flux to the oxidative pentose phosphate pathway and TCA cycle from glucose and increase relative flux to lactate. These results provide the first report of reaction-level carbon fluxes in platelets and allow us to distinguish metabolic fluxes with much higher resolution than previous studies.
Keywords: Blood platelets; Metabolic flux analysis; Metabolomics; Thrombin.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures


References
-
- Abonnenc M, Crettaz D, Marvin L, Grund B, Sonego G, Bardyn M, Tissot J-D, Prudent M, Rochat B, Lion N, 2016. Metabolomic profiling highlights oxidative damages in platelet concentrates treated for pathogen inactivation and shows protective role of urate. Metabolomics 12.
-
- Ahn WS, Antoniewicz MR, 2013. Parallel labeling experiments with [1,2-(13)C]glucose and [U-(13)C]glutamine provide new insights into CHO cell metabolism. Metab Eng 15, 34–47. - PubMed
-
- Aibibula M, Naseem KM, Sturmey RG, 2018. Glucose metabolism and metabolic flexibility in blood platelets. J Thromb Haemost 16, 2300–2314. - PubMed
-
- Antoniewicz MR, 2013. Dynamic metabolic flux analysis--tools for probing transient states of metabolic networks. Curr Opin Biotechnol 24, 973–8. - PubMed
-
- Antoniewicz MR, 2015. Methods and advances in metabolic flux analysis: a mini-review. J Ind Microbiol Biotechnol 42, 317–325. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

