A new insight into sex-specific non-coding RNAs and networks in response to SARS-CoV-2
- PMID: 34954105
- PMCID: PMC8695320
- DOI: 10.1016/j.meegid.2021.105195
A new insight into sex-specific non-coding RNAs and networks in response to SARS-CoV-2
Abstract
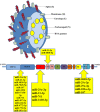
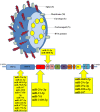
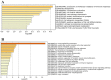
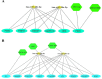
SARS-CoV-2 is the RNA virus responsible for COVID-19, the prognosis of which has been found to be slightly worse in men. The present study aimed to analyze the expression of different mRNAs and their regulatory molecules (miRNAs and lncRNAs) to consider the potential existence of sex-specific expression patterns and COVID-19 susceptibility using bioinformatics analysis. The binding sites of all human mature miRNA sequences on the SARS-CoV-2 genome nucleotide sequence were predicted by the miRanda tool. Sequencing data was excavated using the Galaxy web server from GSE157103, and the output of feature counts was analyzed using DEseq2 packages to obtain differentially expressed genes (DEGs). Gene set enrichment analysis (GSEA) and DEG annotation analyses were performed using the ToppGene and Metascape tools. Using the RNA Interactome Database, we predicted interactions between differentially expressed lncRNAs and differentially expressed mRNAs. Finally, their networks were constructed with top miRNAs. We identified 11 miRNAs with three to five binding sites on the SARS-COVID-2 genome reference. MiR-29c-3p, miR-21-3p, and miR-6838-5p occupied four binding sites, and miR-29a-3p had five binding sites on the SARS-CoV-2 genome. Moreover, miR-29a-3p, and miR-29c-3p were the top miRNAs targeting DEGs. The expression levels of miRNAs (125, 181b, 130a, 29a, b, c, 212, 181a, 133a) changed in males with COVID-19, in whom they regulated ACE2 expression and affected the immune response by affecting phagosomes, complement activation, and cell-matrix adhesion. Our results indicated that XIST lncRNA was up-regulated, and TTTY14, TTTY10, and ZFY-AS1 lncRN as were down-regulated in both ICU and non-ICU men with COVID-19. Dysregulation of noncoding-RNAs has critical effects on the pathophysiology of men with COVID-19, which is why they may be used as biomarkers and therapeutic agents. Overall, our results indicated that the miR-29 family target regulation patterns and might become promising biomarkers for severity and survival outcome in men with COVID-19.
Keywords: COVID-19; lncRNA; miR-29 family; microRNA.
Copyright © 2021. Published by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Computational Analysis of Targeting SARS-CoV-2, Viral Entry Proteins ACE2 and TMPRSS2, and Interferon Genes by Host MicroRNAs.Genes (Basel). 2020 Nov 16;11(11):1354. doi: 10.3390/genes11111354. Genes (Basel). 2020. PMID: 33207533 Free PMC article.
-
The role of microRNAs in modulating SARS-CoV-2 infection in human cells: a systematic review.Infect Genet Evol. 2021 Jul;91:104832. doi: 10.1016/j.meegid.2021.104832. Epub 2021 Apr 1. Infect Genet Evol. 2021. PMID: 33812037 Free PMC article.
-
In Silico Identification of miRNA-lncRNA Interactions in Male Reproductive Disorder Associated with COVID-19 Infection.Cells. 2021 Jun 12;10(6):1480. doi: 10.3390/cells10061480. Cells. 2021. PMID: 34204705 Free PMC article.
-
SARS-CoV-2 structural coverage map reveals viral protein assembly, mimicry, and hijacking mechanisms.Mol Syst Biol. 2021 Sep;17(9):e10079. doi: 10.15252/msb.202010079. Mol Syst Biol. 2021. PMID: 34519429 Free PMC article.
-
Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection.J Biol Chem. 2021 Jan-Jun;296:100135. doi: 10.1074/jbc.REV120.015980. Epub 2020 Dec 6. J Biol Chem. 2021. PMID: 33268377 Free PMC article. Review.
Cited by
-
Analyzing the expression pattern of the noncoding RNAs (HOTAIR, PVT-1, XIST, H19, and miRNA-34a) in PBMC samples of patients with COVID-19, according to the disease severity in Iran during 2022-2023: A cross-sectional study.Health Sci Rep. 2024 Feb 7;7(2):e1861. doi: 10.1002/hsr2.1861. eCollection 2024 Feb. Health Sci Rep. 2024. PMID: 38332929 Free PMC article.
-
ceRNA network construction and identification of hub genes as novel therapeutic targets for age-related cataracts using bioinformatics.PeerJ. 2023 Mar 23;11:e15054. doi: 10.7717/peerj.15054. eCollection 2023. PeerJ. 2023. PMID: 36987450 Free PMC article.
-
COVID-19 in patients with anemia and haematological malignancies: risk factors, clinical guidelines, and emerging therapeutic approaches.Cell Commun Signal. 2024 Feb 15;22(1):126. doi: 10.1186/s12964-023-01316-9. Cell Commun Signal. 2024. PMID: 38360719 Free PMC article. Review.
-
Unveiling the Hidden Regulators: The Impact of lncRNAs on Zoonoses.Int J Mol Sci. 2024 Mar 21;25(6):3539. doi: 10.3390/ijms25063539. Int J Mol Sci. 2024. PMID: 38542512 Free PMC article. Review.
-
Genetic and Epigenetic Intersections in COVID-19-Associated Cardiovascular Disease: Emerging Insights and Future Directions.Biomedicines. 2025 Feb 16;13(2):485. doi: 10.3390/biomedicines13020485. Biomedicines. 2025. PMID: 40002898 Free PMC article. Review.
References
-
- Arisan E.D., Dart A., Grant G.H., Arisan S., Cuhadaroglu S., Lange S., Uysal-Onganer P. The prediction of miRNAs in SARS-CoV-2 genomes: hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. 2020;12(6):614. Jun. - PMC - PubMed
-
- Brown C.J., Ballabio A., Rupert J.L., Lafreniere R.G., Grompe M., Tonlorenzi R., Willard H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. Jan. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

