A β-Catenin-TCF-Sensitive Locus Control Region Mediates GUCY2C Ligand Loss in Colorectal Cancer
- PMID: 34954189
- PMCID: PMC9073733
- DOI: 10.1016/j.jcmgh.2021.12.014
A β-Catenin-TCF-Sensitive Locus Control Region Mediates GUCY2C Ligand Loss in Colorectal Cancer
Abstract
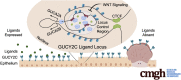
Background & aims: Sporadic colorectal cancers arise from initiating mutations in APC, producing oncogenic β-catenin/TCF-dependent transcriptional reprogramming. Similarly, the tumor suppressor axis regulated by the intestinal epithelial receptor GUCY2C is among the earliest pathways silenced in tumorigenesis. Retention of the receptor, but loss of its paracrine ligands, guanylin and uroguanylin, is an evolutionarily conserved feature of colorectal tumors, arising in the earliest dysplastic lesions. Here, we examined a mechanism of GUCY2C ligand transcriptional silencing by β-catenin/TCF signaling.
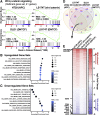
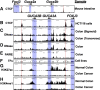
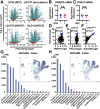
Methods: We performed RNA sequencing analysis of 4 unique conditional human colon cancer cell models of β-catenin/TCF signaling to map the core Wnt-transcriptional program. We then performed a comparative analysis of orthogonal approaches, including luciferase reporters, chromatin immunoprecipitation sequencing, CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats) knockout, and CRISPR epigenome editing, which were cross-validated with human tissue chromatin immunoprecipitation sequencing datasets, to identify functional gene enhancers mediating GUCY2C ligand loss.
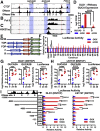
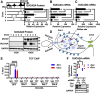
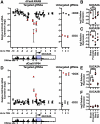
Results: RNA sequencing analyses reveal the GUCY2C hormones as 2 of the most sensitive targets of β-catenin/TCF signaling, reflecting transcriptional repression. The GUCY2C hormones share an insulated genomic locus containing a novel locus control region upstream of the guanylin promoter that mediates the coordinated silencing of both genes. Targeting this region with CRISPR epigenome editing reconstituted GUCY2C ligand expression, overcoming gene inactivation by mutant β-catenin/TCF signaling.
Conclusions: These studies reveal DNA elements regulating corepression of GUCY2C ligand transcription by β-catenin/TCF signaling, reflecting a novel pathophysiological step in tumorigenesis. They offer unique genomic strategies that could reestablish hormone expression in the context of canonical oncogenic mutations to reconstitute the GUCY2C axis and oppose transformation.
Keywords: Guanylin; Wnt signaling; super-enhancer; uroguanylin.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

