Evidence for a mouse origin of the SARS-CoV-2 Omicron variant
- PMID: 34954396
- PMCID: PMC8702434
- DOI: 10.1016/j.jgg.2021.12.003
Evidence for a mouse origin of the SARS-CoV-2 Omicron variant
Abstract
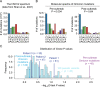
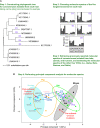
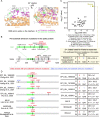
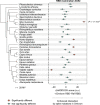
The rapid accumulation of mutations in the SARS-CoV-2 Omicron variant that enabled its outbreak raises questions as to whether its proximal origin occurred in humans or another mammalian host. Here, we identified 45 point mutations that Omicron acquired since divergence from the B.1.1 lineage. We found that the Omicron spike protein sequence was subjected to stronger positive selection than that of any reported SARS-CoV-2 variants known to evolve persistently in human hosts, suggesting a possibility of host-jumping. The molecular spectrum of mutations (i.e., the relative frequency of the 12 types of base substitutions) acquired by the progenitor of Omicron was significantly different from the spectrum for viruses that evolved in human patients but resembled the spectra associated with virus evolution in a mouse cellular environment. Furthermore, mutations in the Omicron spike protein significantly overlapped with SARS-CoV-2 mutations known to promote adaptation to mouse hosts, particularly through enhanced spike protein binding affinity for the mouse cell entry receptor. Collectively, our results suggest that the progenitor of Omicron jumped from humans to mice, rapidly accumulated mutations conducive to infecting that host, then jumped back into humans, indicating an inter-species evolutionary trajectory for the Omicron outbreak.
Keywords: Evolutionary origins; Molecular spectrum of mutations; Omicron; Receptor-binding domain; SARS-CoV-2; Spike-ACE2 interaction.
Copyright © 2021 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

