Genome-Wide Analysis of Simple Sequence Repeats in Cabbage (Brassica oleracea L.)
- PMID: 34956251
- PMCID: PMC8695497
- DOI: 10.3389/fpls.2021.726084
Genome-Wide Analysis of Simple Sequence Repeats in Cabbage (Brassica oleracea L.)
Abstract
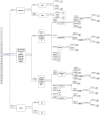
Cabbage (Brassica oleracea L. var. capitata) accounts for a critical vegetable crop belonging to Brassicaceae family, and it has been extensively planted worldwide. Simple sequence repeats (SSRs), the markers with high polymorphism and co-dominance degrees, offer a crucial genetic research resource. The current work identified totally 64,546 perfect and 93,724 imperfect SSR motifs in the genome of the cabbage 'TO1000.' Then, we divided SSRs based on the respective overall length and repeat number into different linkage groups. Later, we characterized cabbage genomes from the perspectives of motif length, motif-type classified and SSR level, and compared them across cruciferous genomes. Furthermore, a large set of 64,546 primer pairs were successfully identified, which generated altogether 1,113 SSR primers, including 916 (82.3%) exhibiting repeated and stable amplification. In addition, there were 32 informative SSR markers screened, which might decide 32 cabbage genotypes for their genetic diversity, with level of polymorphism information of 0.14-0.88. Cultivars were efficiently identified by the new strategy designating manual diagram for identifying cultivars. Lastly, 32 cabbage accessions were clearly separately by five Bol-SSR markers. Besides, we verified whether such SSRs were available and transferable in 10 Brassicaceae relatives. Based on the above findings, those genomic SSR markers identified in the present work may facilitate cabbage research, which lay a certain foundation for further gene tagging and genetic linkage analyses, like marker-assisted selection, genetic mapping, as well as comparative genomic analysis.
Keywords: SSR; cabbage; genetic diversity; genome; manual cultivar identification diagram; molecular makers.
Copyright © 2021 Xu, Xing, Song, Yan, Lu and Zeng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Ahmed H. G. M. D., Kashif M., Rashid M. A. R., Sajjad M., Zeng Y. W. (2020). Genome wide diversity in bread wheat evaluated by SSR markers. Int. J. Agric. Biol. 24, 263–272. doi: 10.17957/IJAB/15.1433 - DOI
LinkOut - more resources
Full Text Sources
