Imaging and visualizing SARS-CoV-2 in a new era for structural biology
- PMID: 34956593
- PMCID: PMC8504884
- DOI: 10.1098/rsfs.2021.0019
Imaging and visualizing SARS-CoV-2 in a new era for structural biology
Abstract
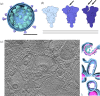
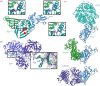
The SARS-CoV-2 pandemic has had a global impact and has put scientific endeavour in the spotlight, perhaps more than any previous viral outbreak. Fortuitously, the pandemic came at a time when decades of research in multiple scientific fields could be rapidly brought to bear, and a new generation of vaccine platforms was on the cusp of clinical maturity. SARS-CoV-2 also emerged at the inflection point of a technological revolution in macromolecular imaging by cryo-electron microscopy, fuelled by a confluence of major technological advances in sample preparation, optics, detectors and image processing software, that complemented pre-existing techniques. Together, these advances enabled us to visualize SARS-CoV-2 and its components more rapidly, in greater detail, and in a wider variety of biologically relevant contexts than would have been possible even a few years earlier. The resulting ultrastructural information on SARS-CoV-2 and how it interacts with the host cell has played a critical role in the much-needed accelerated development of COVID-19 vaccines and therapeutics. Here, we review key imaging modalities used to visualize SARS-CoV-2 and present select example data, which have provided us with an exceptionally detailed picture of this virus.
Keywords: SARS-CoV-2; X-ray crystallography; cryo-electron microscopy; cryo-electron tomography; multiscale imaging; structural biology.
© 2021 The Authors.
Figures



References
-
- Crowther RA (ed.). 2016. The resolution revolution: recent advances in CryoEM, vol. 579, Methods in enzymology. London, UK: Academic Press.
-
- Iwanowski D. 1903. Über die Mosaikkrankheit der Tabakspflanze. Z. Pflanzenkrankh. Pflanzenschutz 13, 1-41.
-
- Marton L. 1934. Electron microscopy of biological objects. Nature 133, 911.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
