A stepwise mutagenesis approach using histidine and acidic amino acid to engineer highly pH-dependent protein switches
- PMID: 34956814
- PMCID: PMC8686790
- DOI: 10.1007/s13205-021-03079-x
A stepwise mutagenesis approach using histidine and acidic amino acid to engineer highly pH-dependent protein switches
Abstract
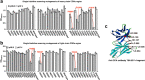
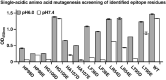
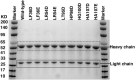
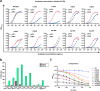
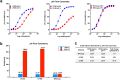
Antibody-based drugs can be highly toxic, because they target normal tissue as well as tumor tissue. The pH value of the extracellular microenvironments around tumor tissues is lower than that of normal tissues. Therefore, antibodies that engage in pH-dependent binding at slightly acidic pH are crucial for improving the safety of antibody-based drugs. Thus, we implemented a stepwise mutagenesis approach to engineering pH-dependent antibodies capable of selective binding in the acidic microenvironment in this study. The first step involved single-residue histidine scanning mutagenesis of the antibody's complementarity-determining regions to prescreen for pH-dependent mutants and identify ionizable sensitive hot-spot residues that could be substituted by acidic amino acids to obtain pH-dependent antibodies. The second step involved single-acidic amino acid residue substitutions of the identified residues and the assessment of pH-dependent binding. We identified six ionizable sensitive hot-spot residues using single-histidine scanning mutagenesis. Nine pH-dependent antibodies were isolated using single-acidic amino acid residue mutagenesis at the six hot-spot residue positions. Relative to wild-type anti-CEA chimera antibody, the binding selectivity of the best performing mutant was improved by approximately 32-fold according to ELISA and by tenfold according to FACS assay. The mutant had a high affinity in the pH range of 5.5-6.0. This study supports the development of pH-dependent protein switches and increases our understanding of the role of ionizable residues in protein interfaces. The stepwise mutagenesis approach is rapid, general, and robust and is expected to produce pH-sensitive protein affinity reagents for various applications.
Keywords: Antibody engineering; Carcinoembryonic antigen; Hot-spot residues; Tumor microenvironments.
© King Abdulaziz City for Science and Technology 2021.
Conflict of interest statement
Conflict of interestThe authors declare no conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

