Copy Number Alterations in Hepatoblastoma: Literature Review and a Brazilian Cohort Analysis Highlight New Biological Pathways
- PMID: 34956867
- PMCID: PMC8692715
- DOI: 10.3389/fonc.2021.741526
Copy Number Alterations in Hepatoblastoma: Literature Review and a Brazilian Cohort Analysis Highlight New Biological Pathways
Abstract
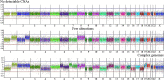
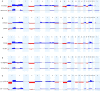
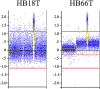
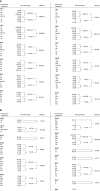
Hepatoblastoma (HB) is a rare embryonal tumor, although it is the most common pediatric liver cancer. The aim of this study was to provide an accurate cytogenomic profile of this type of cancer, for which information in cancer databases is lacking. We performed an extensive literature review of cytogenetic studies on HBs disclosing that the most frequent copy number alterations (CNAs) are gains of 1q, 2/2q, 8/8q, and 20; and losses at 1p and 4q. Furthermore, the CNA profile of a Brazilian cohort of 26 HBs was obtained by array-CGH; the most recurrent CNAs were the same as shown in the literature review. Importantly, HBs from female patients, high-risk stratification tumors, tumors who developed in older patients (> 3 years at diagnosis) or from patients with metastasis and/or deceased carried a higher diversity of chromosomal alterations, specifically chromosomal losses at 1p, 4, 11q and 18q. In addition, we distinguished three major CNA profiles: no detectable CNA, few CNAs and tumors with complex genomes. Tumors with simpler genomes exhibited a significant association with the epithelial fetal subtype of HBs; in contrast, the complex genome group included three cases with epithelial embryonal histology, as well as the only HB with HCC features. A significant association of complex HB genomes was observed with older patients who developed high-risk tumors, metastasis, and deceased. Moreover, two patients with HBs exhibiting complex genomes were born with congenital anomalies. Together, these findings suggest that a high load of CNAs, mainly chromosomal losses, particularly losses at 1p and 18, increases the tendency to HB aggressiveness. Additionally, we identified six hot-spot chromosome regions most frequently affected in the entire group: 1q31.3q42.3, 2q23.3q37.3, and 20p13p11.1 gains, besides a 5,3 Mb amplification at 2q24.2q24.3, and losses at 1p36.33p35.1, 4p14 and 4q21.22q25. An in-silico analysis using the genes mapped to these six regions revealed several enriched biological pathways such as ERK Signaling, MicroRNAs in Cancer, and the PI3K-Akt Signaling, in addition to the WNT Signaling pathway; further investigation is required to evaluate if disturbances of these pathways can contribute to HB tumorigenesis. The analyzed gene set was found to be associated with neoplasms, abnormalities of metabolism/homeostasis and liver morphology, as well as abnormal embryonic development and cytokine secretion. In conclusion, we have provided a comprehensive characterization of the spectrum of chromosomal alterations reported in HBs and identified specific genomic regions recurrently altered in a Brazilian HB group, pointing to new biological pathways, and relevant clinical associations.
Keywords: array-CGH; copy number alteration; cytogenomics; hepatoblastoma; pediatric cancer.
Copyright © 2021 Barros, Aguiar, Costa, Rivas, Cypriano, Toledo, Novak, Odone, Cristofani, Carraro, werneck da Cunha, Costa, Vianna-Morgante, Rosenberg and Krepischi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

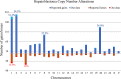
References
-
- Aguiar TFM, Carneiro TN, da Costa CML, Rosenberg C, da Cunha IW, Krepischi ACV. The Genetic and Epigenetic Landscapes of Hepatoblastomas. Appl Cancer Res (2017) 37(1):1–7. doi: 10.1186/s41241-017-0021-0 - DOI
-
- Venkatramani R, Spector LG, Georgieff M, Tomlinson G, Krailo M, Malogolowkin M, et al. Congenital Abnormalities and Hepatoblastoma: A Report from the Children’s Oncology Group (COG) and the Utah Population Database (UPDB). Am J Med Genet Part A (2014) 23(1):1–7. doi: 10.1002/ajmg.a.36638 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

