Aptamer Sandwich Assay for the Detection of SARS-CoV-2 Spike Protein Antigen
- PMID: 34957366
- PMCID: PMC8691202
- DOI: 10.1021/acsomega.1c05521
Aptamer Sandwich Assay for the Detection of SARS-CoV-2 Spike Protein Antigen
Abstract
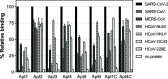
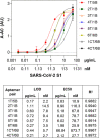
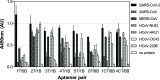
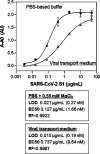
The novel severe acute respiratory syndrome coronavirus (SARS-CoV-2) emerged at the end of 2019, resulting in the ongoing COVID-19 pandemic. The high transmissibility of the virus and the substantial number of asymptomatic individuals have led to an exponential rise in infections worldwide, urgently requiring global containment strategies. Reverse transcription-polymerase chain reaction is the gold standard for the detection of SARS-CoV-2 infections. Antigen tests, targeting the spike (S) or nucleocapsid (N) viral proteins, are considered as complementary tools. Despite their shortcomings in terms of sensitivity and specificity, antigen tests could be deployed for the detection of potentially contagious individuals with high viral loads. In this work, we sought to develop a sandwich aptamer-based assay for the detection of the S protein of SARS-CoV-2. A detailed study on the binding properties of aptamers to the receptor-binding domain of the S protein in search of aptamer pairs forming a sandwich is presented. Screening of aptamer pairs and optimization of assay conditions led to the development of a laboratory-based sandwich assay able to detect 21 ng/mL (270 pM) of the protein with negligible cross-reactivity with the other known human coronaviruses. The detection of 375 pg of the protein in viral transport medium demonstrates the compatibility of the assay with clinical specimens. Finally, successful detection of the S antigen in nasopharyngeal swab samples collected from suspected patients further establishes the suitability of the assay for screening purposes as a complementary tool to assist in the control of the pandemic.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Gorbalenya A. E.; Baker S. C.; Baric R. S.; de Groot R. J.; Drosten C.; Gulyaeva A. A.; Haagmans B. L.; Lauber C.; Leontovich A. M.; Neuman B. W.; Penzar D.; Perlman S.; Poon L. L. M.; Samborskiy D. V.; Sidorov I. A.; Sola I.; Ziebuhr J. The species Severe Acute Respiratory Syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. 10.1038/s41564-020-0695-z. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

