Evaluation of the Suitability of RNAscope as a Technique to Measure Gene Expression in Clinical Diagnostics: A Systematic Review
- PMID: 34957535
- PMCID: PMC8710359
- DOI: 10.1007/s40291-021-00570-2
Evaluation of the Suitability of RNAscope as a Technique to Measure Gene Expression in Clinical Diagnostics: A Systematic Review
Abstract
Objective: To evaluate the application of RNAscope in the clinical diagnostic field compared to the current 'gold standard' methods employed for testing gene expression levels, including immunohistochemistry (IHC), quantitative real time PCR (qPCR), and quantitative reverse transcriptase PCR (qRT-PCR), and to detect genes, including DNA in situ hybridisation (DNA ISH).
Methods: This systematic review searched CINAHL, Medline, Embase and Web of Science databases for studies that were conducted after 2012 and that compared RNAscope with one or more of the 'gold standard' techniques in human samples. QUADAS-2 test was used for the evaluation of the articles' risk of bias. The results were reviewed narratively and analysed qualitatively.
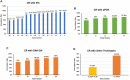
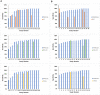
Results: A total of 27 articles (all retrospective studies) were obtained and reviewed. The 27 articles showed a range of low to middle risk of bias scores, as assessed by QUADAS-2 test. 26 articles studied RNAscope within cancer samples. RNAscope was compared to different techniques throughout the included studies (IHC, qPCR, qRT-PCR and DNA ISH). The results confirmed that RNAscope is a highly sensitive and specific method that has a high concordance rate (CR) with qPCR, qRT-PCR, and DNA ISH (81.8-100%). However, the CR with IHC was lower than expected (58.7-95.3%), which is mostly due to the different products that each technique measures (RNA vs. protein).
Discussion: This is the first systematic review to be conducted on the use of RNAscope in the clinical diagnostic field. RNAscope was found to be a reliable and robust method that could complement gold standard techniques currently used in clinical diagnostics to measure gene expression levels or for gene detection. However, there were not enough data to suggest that RNAscope could stand alone in the clinical diagnostic setting, indicating further prospective studies to validate diagnostic accuracy values, in keeping with relevant regulations, followed by cost evaluation are required.
© 2021. The Author(s), under exclusive licence to Springer Nature Switzerland AG.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures