Role of lncRNA LIPE-AS1 in adipogenesis
- PMID: 34957921
- PMCID: PMC8726699
- DOI: 10.1080/21623945.2021.2013415
Role of lncRNA LIPE-AS1 in adipogenesis
Abstract
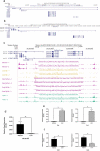
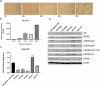
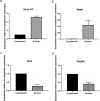
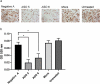
Recent studies have identified long non-coding RNAs (lncRNAs) as potential regulators of adipogenesis. In this study, we have characterized a lncRNA, LIPE-AS1, that spans genes CEACAM1 to LIPE in man with conservation of genomic organization and tissue expression between mouse and man. Tissue-specific expression of isoforms of the murine lncRNA were found in liver and adipose tissue, one of which, designated mLas-V3, overlapped the Lipe gene encoding hormone-sensitive lipase in both mouse and man suggesting that it may have a functional role in adipose tissue. Knock down of expression of mLas-V3 using anti-sense oligos (ASOs) led to a significant decrease in the differentiation of the OP9 pre-adipocyte cell line through the down regulation of the major adipogenic transcription factors Pparg and Cebpa. Knock down of mLas-V3 induced apoptosis during the differentiation of OP9 cells as shown by expression of active caspase-3, a change in the localization of LIP/LAP isoforms of C/EBPβ, and expression of the cellular stress induced factors CHOP, p53, PUMA, and NOXA. We conclude that mLas-V3 may play a role in protecting against stress associated with adipogenesis, and its absence leads to apoptosis.
Keywords: Ceacam1; LIPE; adipogenesis; apoptosis; long non-coding RNA.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

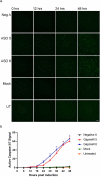

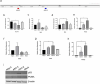
References
-
- Hales CMCM, Fryar CD, Ogden CL.. Prevalence of obesity and severe obesity amound adults: United States, 2017-2018. NCHS data brief, no 360. Hyattsville MD: National Center for Health Statistics; 2020. - PubMed
-
- Prevention CfDCa . National diabetes statistics report. Atlanta GA: Centers for Disease Control and Prevention, U.S. Dept of Health and Human Services; 2020.
-
- Camp HS, Ren D, Leff T. Adipogenesis and fat-cell function in obesity and diabetes. Trends Mol Med. 2002. Sep;8(9):442–447. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
