Structure and function of the ecto-nucleotide pyrophosphatase/phosphodiesterase (ENPP) family: Tidying up diversity
- PMID: 34958798
- PMCID: PMC8808174
- DOI: 10.1016/j.jbc.2021.101526
Structure and function of the ecto-nucleotide pyrophosphatase/phosphodiesterase (ENPP) family: Tidying up diversity
Abstract
Ecto-nucleotide pyrophosphatase/phosphodiesterase (ENPP) family members (ENPP1-7) have been implicated in key biological and pathophysiological processes, including nucleotide and phospholipid signaling, bone mineralization, fibrotic diseases, and tumor-associated immune cell infiltration. ENPPs are single-pass transmembrane ecto-enzymes, with notable exceptions of ENPP2 (Autotaxin) and ENNP6, which are secreted and glycosylphosphatidylinositol (GPI)-anchored, respectively. ENNP1 and ENNP2 are the best characterized and functionally the most interesting members. Here, we review the structural features of ENPP1-7 to understand how they evolved to accommodate specific substrates and mediate different biological activities. ENPPs are defined by a conserved phosphodiesterase (PDE) domain. In ENPP1-3, the PDE domain is flanked by two N-terminal somatomedin B-like domains and a C-terminal inactive nuclease domain that confers structural stability, whereas ENPP4-7 only possess the PDE domain. Structural differences in the substrate-binding site endow each protein with unique characteristics. Thus, ENPP1, ENPP3, ENPP4, and ENPP5 hydrolyze nucleotides, whereas ENPP2, ENPP6, and ENNP7 evolved as phospholipases through adaptions in the catalytic domain. These adaptations explain the different biological and pathophysiological functions of individual members. Understanding the ENPP members as a whole advances our insights into common mechanisms, highlights their functional diversity, and helps to explore new biological roles.
Keywords: autotaxin; cancer; drug development; mineralization; phosphodiesterases; phospholipase; pyrophosphate; signaling; structure–function.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

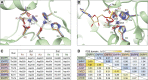

References
-
- van Driel I.R., Goding J.W. Plasma cell membrane glycoprotein PC-1. Primary structure deduced from cDNA clones. J. Biol. Chem. 1987;262:4882–4887. - PubMed
-
- Goding J.W., Terkeltaub R., Maurice M., Deterre P., Sali A., Belli S.I. Ecto-phosphodiesterase/pyrophosphatase of lymphocytes and non-lymphoid cells: Structure and function of the PC-1 family. Immunol. Rev. 1998;161:11–26. - PubMed
-
- Bollen M., Gijsbers R., Ceulemans H., Stalmans W., Stefan C. Nucleotide pyrophosphatases/phosphodiesterases on the move. Crit. Rev. Biochem. Mol. Biol. 2000;35:393–432. - PubMed
-
- Tokumura A., Majima E., Kariya Y., Tominaga K., Kogure K., Yasuda K., Fukuzawa K. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J. Biol. Chem. 2002;277:39436–39442. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

