Genome-wide analysis of the response to ivermectin treatment by a Swedish field population of Haemonchus contortus
- PMID: 34959200
- PMCID: PMC8718930
- DOI: 10.1016/j.ijpddr.2021.12.002
Genome-wide analysis of the response to ivermectin treatment by a Swedish field population of Haemonchus contortus
Abstract
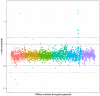
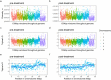
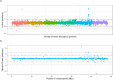
Haemonchus contortus is a pathogenic gastrointestinal nematode of small ruminants and, in part due to its capacity to develop resistance to drugs, contributes to significant losses in the animal production sector worldwide. Despite decades of research, comparatively little is known about the specific mechanism(s) driving resistance to drugs such as ivermectin in this species. Here we describe a genome-wide approach to detect evidence of selection by ivermectin treatment in a field population of H. contortus from Sweden, using parasites sampled from the same animals before and seven days after ivermectin exposure followed by whole-genome sequencing. Despite an 89% reduction in parasites recovered after treatment measured by the fecal egg count reduction test, the surviving population was highly genetically similar to the population before treatment, suggesting that resistance has likely evolved over time and that resistance alleles are present on diverse haplotypes. Pairwise gene and SNP frequency comparisons indicated the highest degree of differentiation was found at the terminal end of chromosome 4, whereas the most striking difference in nucleotide diversity was observed in a region on chromosome 5 previously reported to harbor a major quantitative trait locus involved in ivermectin resistance. These data provide novel insight into the genome-wide effect of ivermectin selection in a field population as well as confirm the importance of the previously established quantitative trait locus in the development of resistance to ivermectin.
Keywords: Anthelmintic resistance; Haemonchus contortus; Ivermectin; Pool-seq; Whole-genome sequencing.
Copyright © 2021 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
The authors of this manuscript certify that they have NO affiliations with or involvement in any organization or entity with any financial interest, or non-financial interest in the subject matter discussed in this manuscript.
Figures

References
-
- Abkallo H.M., Martinelli A., Inoue M., Ramaprasad A., Xangsayarath P., Gitaka J., Tang J., Yahata K., Zoungrana A., Mitaka H., Acharjee A., Datta P.P., Hunt P., Carter R., Kaneko O., Mustonen V., Illingworth C.J.R., Pain A., Culleton R. Rapid identification of genes controlling virulence and immunity in malaria parasites. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006447. - DOI - PMC - PubMed
-
- Cazajous T., Prevot F., Kerbiriou A., Milhes M., Grisez C., Tropee A., Godart C., Aragon A., Jacquiet P. Multiple-resistance to ivermectin and benzimidazole of a Haemonchus contortus population in a sheep flock from mainland France, first report. Vet. Parasitol. Reg. Stud. Reports. 2018;14:103–105. doi: 10.1016/j.vprsr.2018.09.005. - DOI - PubMed
-
- Charlier J., Rinaldi L., Musella V., Ploeger H.W., Chartier C., Vineer H.R., Hinney B., von Samson-Himmelstjerna G., Băcescu B., Mickiewicz M., Mateus T.L., Martinez-Valladares M., Quealy S., Azaizeh H., Sekovska B., Akkari H., Petkevicius S., Hektoen L., Höglund J., Morgan E.R., Bartley D.J., Claerebout E. Initial assessment of the economic burden of major parasitic helminth infections to the ruminant livestock industry in Europe. Prev. Vet. Med. 2020;182:105103. doi: 10.1016/j.prevetmed.2020.105103. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

