Host Range, Biology, and Species Specificity of Seven-Segmented Influenza Viruses-A Comparative Review on Influenza C and D
- PMID: 34959538
- PMCID: PMC8704295
- DOI: 10.3390/pathogens10121583
Host Range, Biology, and Species Specificity of Seven-Segmented Influenza Viruses-A Comparative Review on Influenza C and D
Abstract
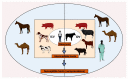
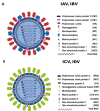
Other than genome structure, influenza C (ICV), and D (IDV) viruses with seven-segmented genomes are biologically different from the eight-segmented influenza A (IAV), and B (IBV) viruses concerning the presence of hemagglutinin-esterase fusion protein, which combines the function of hemagglutinin and neuraminidase responsible for receptor-binding, fusion, and receptor-destroying enzymatic activities, respectively. Whereas ICV with humans as primary hosts emerged nearly 74 years ago, IDV, a distant relative of ICV, was isolated in 2011, with bovines as the primary host. Despite its initial emergence in swine, IDV has turned out to be a transboundary bovine pathogen and a broader host range, similar to influenza A viruses (IAV). The receptor specificities of ICV and IDV determine the host range and the species specificity. The recent findings of the presence of the IDV genome in the human respiratory sample, and high traffic human environments indicate its public health significance. Conversely, the presence of ICV in pigs and cattle also raises the possibility of gene segment interactions/virus reassortment between ICV and IDV where these viruses co-exist. This review is a holistic approach to discuss the ecology of seven-segmented influenza viruses by focusing on what is known so far on the host range, seroepidemiology, biology, receptor, phylodynamics, species specificity, and cross-species transmission of the ICV and IDV.
Keywords: biology; eight-segmented; host range; influenza C; influenza D; receptor 9; seven-segmented; species specificity; structure; transmission.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Fields Virology: Emerging Viruses. Volume 1 Wolters Kluwer; Philadelphia, PA, USA: 2021.
-
- Kawano J., Onta T., Kida H., Yanagawa R. Distribution of antibodies in animals against influenza B and C viruses. Jpn. J. Vet. Res. 1978;26:74–80. - PubMed
-
- Chang C.P., New A.E., Taylor J.F., Chiang H.S. Influenza virus isolations from dogs during a human epidemic in Taiwan. Int. J. Zoonoses. 1976;3:61–64. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

