Comprehensive Transcriptome and Pathway Analyses Revealed Central Role for Fascin in Promoting Triple-Negative Breast Cancer Progression
- PMID: 34959629
- PMCID: PMC8708558
- DOI: 10.3390/ph14121228
Comprehensive Transcriptome and Pathway Analyses Revealed Central Role for Fascin in Promoting Triple-Negative Breast Cancer Progression
Abstract
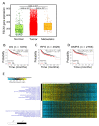
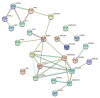
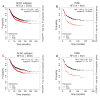
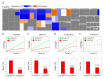
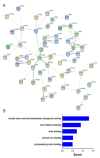
Recent years have witnessed major progress in development of novel therapeutic agents such as chemotherapy, targeted therapy and immune checkpoint inhibitors for breast cancer. However, cancer-related death remains high especially in triple-negative breast cancer (TNBC) due limited therapeutic options. Development of targeted therapies for TNBC requires better understanding of biology and signaling networks that promote disease progression. Fascin, an actin bundling protein, was identified as a key regulator of many signaling pathways that contribute to breast cancer progression. Herein, fascin ShRNA was used to generate stable fascin knockdown (FSCN1KD) in the MDA-MB-231 TNBC cell line and then were subjected to comprehensive mRNA and miRNA transcriptome analysis. We identified 129 upregulated and 114 downregulated mRNA transcripts, while 14 miRNAs were differentially expressed in FSCN1KD. Ingenuity pathway analysis (IPA) was used to predict the impact of differentially expressed transcripts on signaling pathways and functional categories and to construct miRNA-mRNA regulatory networks in the context of FSCN1 knockdown. Compared to FSCN1KD, fascin-positive (FSCN1CON) breast cancer cells showed enrichment in genes promoting cellular proliferation, migration, survival, DNA replication and repair. Expression of FSCN1high (identified in BRCA dataset from TCGA) in conjunction with elevated expression of the top 10 upregulated or decreased expression of the top 10 downregulated genes (identified in our FSCN1CON vs. FSCN1KD) correlates with worst survival outcome. Taken together, these data confirmed fascin's role in promoting TNBC progression, and identified a novel opportunity for therapeutic interventions via targeting those FSCN1-related transcripts.
Keywords: IPA; breast cancer; fascin; miRNA; pathway analysis; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Balduzzi A., Bagnardi V., Rotmensz N., Dellapasqua S., Montagna E., Cardillo A., Viale G., Veronesi P., Intra M., Luini A., et al. Survival Outcomes in Breast Cancer Patients with Low Estrogen/Progesterone Receptor Expression. Clin. Breast Cancer. 2013;14:258–264. doi: 10.1016/j.clbc.2013.10.019. - DOI - PubMed
-
- Dellapasqua S., Bagnardi V., Balduzzi A., Iorfida M., Rotmensz N., Santillo B., Viale G., Ghisini R., Veronesi P., Luini A., et al. Outcomes of patients with breast cancer who present with ipsilateral supraclavicular or internal mammary lymph node metastases. Clin. Breast Cancer. 2014;14:53–60. doi: 10.1016/j.clbc.2013.09.008. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

