Comirnaty-Elicited and Convalescent Sera Recognize Different Spike Epitopes
- PMID: 34960165
- PMCID: PMC8708883
- DOI: 10.3390/vaccines9121419
Comirnaty-Elicited and Convalescent Sera Recognize Different Spike Epitopes
Abstract
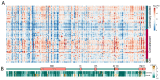
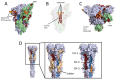
Many of the approved SARS-CoV-2 vaccines are based on a stabilized variant of the spike protein. This raises the question of whether the immune response against the stabilized spike is identical to the immune response that is elicited by the native spike in the case of a SARS-CoV-2 infection. Using a peptide array-based approach, we analysed the binding of antibodies from Comirnaty-elicited, convalescent, and control sera to the peptides covering the spike protein. A total of 37 linear epitopes were identified. A total of 26 of these epitopes were almost exclusively recognized by the convalescent sera. Mapping these epitopes to the spike structures revealed that most of these 26 epitopes are masked in the pre-fusion structure. In particular, in the conserved central helix, three epitopes that are only exposed in the post-fusion conformation were identified. This indicates a higher spike-specific antibody diversity in convalescent sera. These differences could be relevant for the breadth of spike-specific immune response.
Keywords: SARS-CoV-2 spike; convalescent; linear epitopes; peptide array; stabilized spike protein.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., et al. A prefusion SARS-CoV-2 spike RNA vaccine is highly immunogenic and prevents lung infection in non-human primates. biorXiv. 2020 doi: 10.1101/2020.09.08.280818. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

