Fast, Reliable, and Simple Point-of-Care-like Adaptation of RT-qPCR for the Detection of SARS-CoV-2 for Use in Hospital Emergency Departments
- PMID: 34960682
- PMCID: PMC8707628
- DOI: 10.3390/v13122413
Fast, Reliable, and Simple Point-of-Care-like Adaptation of RT-qPCR for the Detection of SARS-CoV-2 for Use in Hospital Emergency Departments
Abstract
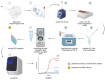
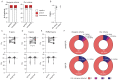
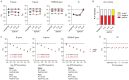
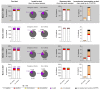
During COVID-19 pandemics, the availability of testing has often been a limiting factor during patient admissions into the hospital. To circumvent this problem, we adapted an existing diagnostic assay, Seegene Allplex SARS-CoV-2, into a point-of-care-style direct qPCR (POC dqPCR) assay and implemented it in the Emergency Department of Clinical Hospital Center Rijeka, Croatia. In a 4-month analysis, we tested over 10,000 patients and demonstrated that POC-dqPCR is robust and reliable and can be successfully implemented in emergency departments and similar near-patient settings and can be performed by medical personnel with little prior experience in qPCR.
Keywords: COVID-19; SARS-CoV-2; direct qPCR; dqPCR; emergency department; molecular diagnostics; point-of-care PCR; qPCR.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

