Interactions between the Nucleoprotein and the Phosphoprotein of Pneumoviruses: Structural Insight for Rational Design of Antivirals
- PMID: 34960719
- PMCID: PMC8706346
- DOI: 10.3390/v13122449
Interactions between the Nucleoprotein and the Phosphoprotein of Pneumoviruses: Structural Insight for Rational Design of Antivirals
Abstract
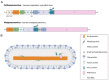
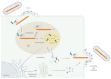
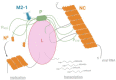
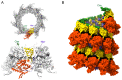
Pneumoviruses include pathogenic human and animal viruses, the most known and studied being the human respiratory syncytial virus (hRSV) and the metapneumovirus (hMPV), which are the major cause of severe acute respiratory tract illness in young children worldwide, and main pathogens infecting elderly and immune-compromised people. The transcription and replication of these viruses take place in specific cytoplasmic inclusions called inclusion bodies (IBs). These activities depend on viral polymerase L, associated with its cofactor phosphoprotein P, for the recognition of the viral RNA genome encapsidated by the nucleoprotein N, forming the nucleocapsid (NC). The polymerase activities rely on diverse transient protein-protein interactions orchestrated by P playing the hub role. Among these interactions, P interacts with the NC to recruit L to the genome. The P protein also plays the role of chaperone to maintain the neosynthesized N monomeric and RNA-free (called N0) before specific encapsidation of the viral genome and antigenome. This review aims at giving an overview of recent structural information obtained for hRSV and hMPV P, N, and more specifically for P-NC and N0-P complexes that pave the way for the rational design of new antivirals against those viruses.
Keywords: HMPV; RSV; antivirals; nucleocapsid; nucleoprotein; phosphoprotein; pneumoviruses; protein-protein interaction; structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Collins P.L., Crowe J.E. Respiratory Syncytial Virus and Metapneumovirus. In: Knipe D.M., Howley P.M., editors. Fields Virology. 5th ed. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007. pp. 1601–1646.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

