Plasma Proteome Fingerprints Reveal Distinctiveness and Clinical Outcome of SARS-CoV-2 Infection
- PMID: 34960725
- PMCID: PMC8706135
- DOI: 10.3390/v13122456
Plasma Proteome Fingerprints Reveal Distinctiveness and Clinical Outcome of SARS-CoV-2 Infection
Abstract
Background: We evaluated how plasma proteomic signatures in patients with suspected COVID-19 can unravel the pathophysiology, and determine kinetics and clinical outcome of the infection.
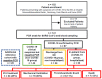
Methods: Plasma samples from patients presenting to the emergency department (ED) with symptoms of COVID-19 were stratified into: (1) patients with suspected COVID-19 that was not confirmed (n = 44); (2) non-hospitalized patients with confirmed COVID-19 (n = 44); (3) hospitalized patients with confirmed COVID-19 (n = 53) with variable outcome; and (4) patients presenting to the ED with minor diseases unrelated to SARS-CoV-2 infection (n = 20). Besides standard of care diagnostics, 177 circulating proteins related to inflammation and cardiovascular disease were analyzed using proximity extension assay (PEA, Olink) technology.
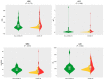
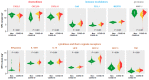
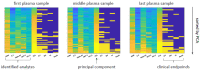
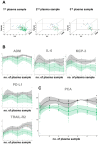
Results: Comparative proteome analysis revealed 14 distinct proteins as highly associated with SARS-CoV-2 infection and 12 proteins with subsequent hospitalization (p < 0.001). ADM, IL-6, MCP-3, TRAIL-R2, and PD-L1 were each predictive for death (AUROC curve 0.80-0.87). The consistent increase of these markers, from hospital admission to intensive care and fatality, supported the concept that these proteins are of major clinical relevance.
Conclusions: We identified distinct plasma proteins linked to the presence and course of COVID-19. These plasma proteomic findings may translate to a protein fingerprint, helping to assist clinical management decisions.
Keywords: COVID-19; emergency medicine; proteomics; proximity extension assay; risk prediction.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures

References
-
- Wynants L., Van Calster B., Collins G.S., Riley R.D., Heinze G., Schuit E., Bonten M.M.J., Dahly D.L., Damen J.A.A., Debray T.P.A., et al. Prediction Models for Diagnosis and Prognosis of Covid-19 Infection: Systematic Review and Critical Appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. - DOI - PMC - PubMed
-
- Goodman K.E., Magder L.S., Baghdadi J.D., Pineles L., Levine A.R., Perencevich E.N., Harris A.D. Impact of Sex and Metabolic Comorbidities on Covid-19 Mortality Risk across Age Groups: 66,646 Inpatients across 613 U.S. Hospitals. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1787. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

