Extensive loss of Wnt genes in Tardigrada
- PMID: 34961481
- PMCID: PMC8711157
- DOI: 10.1186/s12862-021-01954-y
Extensive loss of Wnt genes in Tardigrada
Abstract
Background: Wnt genes code for ligands that activate signaling pathways during development in Metazoa. Through the canonical Wnt (cWnt) signaling pathway, these genes regulate important processes in bilaterian development, such as establishing the anteroposterior axis and posterior growth. In Arthropoda, Wnt ligands also regulate segment polarity, and outgrowth and patterning of developing appendages. Arthropods are part of a lineage called Panarthropoda that includes Onychophora and Tardigrada. Previous studies revealed potential roles of Wnt genes in regulating posterior growth, segment polarity, and growth and patterning of legs in Onychophora. Unlike most other panarthropods, tardigrades lack posterior growth, but retain segmentation and appendages. Here, we investigated Wnt genes in tardigrades to gain insight into potential roles that these genes play during development of the highly compact and miniaturized tardigrade body plan.
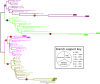
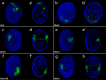
Results: We analyzed published genomes for two representatives of Tardigrada, Hypsibius exemplaris and Ramazzottius varieornatus. We identified single orthologs of Wnt4, Wnt5, Wnt9, Wnt11, and WntA, as well as two Wnt16 paralogs in both tardigrade genomes. We only found a Wnt2 ortholog in H. exemplaris. We could not identify orthologs of Wnt1, Wnt6, Wnt7, Wnt8, or Wnt10. We identified most other components of cWnt signaling in both tardigrade genomes. However, we were unable to identify an ortholog of arrow/Lrp5/6, a gene that codes for a Frizzled co-receptor of Wnt ligands. Additionally, we found that some other animals that have lost several Wnt genes and are secondarily miniaturized, like tardigrades, are also missing an ortholog of arrow/Lrp5/6. We analyzed the embryonic expression patterns of Wnt genes in H. exemplaris during developmental stages that span the establishment of the AP axis through segmentation and leg development. We detected expression of all Wnt genes in H. exemplaris besides one of the Wnt16 paralogs. During embryo elongation, expression of several Wnt genes was restricted to the posterior pole or a region between the anterior and posterior poles. Wnt genes were expressed in distinct patterns during segmentation and development of legs in H. exemplaris, rather than in broadly overlapping patterns.
Conclusions: Our results indicate that Wnt signaling has been highly modified in Tardigrada. While most components of cWnt signaling are conserved in tardigrades, we conclude that tardigrades have lost Wnt1, Wnt6, Wnt7, Wnt8, and Wnt10, along with arrow/Lrp5/6. Our expression data may indicate a conserved role of Wnt genes in specifying posterior identities during establishment of the AP axis. However, the loss of several Wnt genes and the distinct expression patterns of Wnt genes during segmentation and leg development may indicate that combinatorial interactions among Wnt genes are less important during tardigrade development compared to many other animals. Based on our results, and comparisons to previous studies, we speculate that the loss of several Wnt genes in Tardigrada may be related to a reduced number of cells and simplified development that accompanied miniaturization and anatomical simplification in this lineage.
Keywords: Miniaturization; Panarthropoda; Posterior growth; Tardigrada; Wnt signaling.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures








References
-
- Liu J, Xu F, Ji P, Li L, Zhang G. Evolutionary dynamics of the Wnt gene family: implications for lophotrochozoans. J Oceanol Limnol. 2018;36:1720–1730.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
