Prognostic biomarker study in patients with clinical stage I esophageal squamous cell carcinoma: JCOG0502-A1
- PMID: 34962019
- PMCID: PMC8898710
- DOI: 10.1111/cas.15251
Prognostic biomarker study in patients with clinical stage I esophageal squamous cell carcinoma: JCOG0502-A1
Abstract
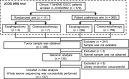
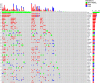
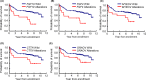
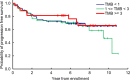
We undertook genomic analyses of Japanese patients with stage I esophageal squamous cell carcinoma (ESCC) to investigate the frequency of genomic alterations and the association with survival outcomes. Biomarker analysis was carried out for patients with clinical stage T1bN0M0 ESCC enrolled in JCOG0502 (UMIN000000551). Whole-exome sequencing (WES) was performed using DNA extracted from formalin-fixed, paraffin-embedded tissue of ESCC and normal tissue or blood sample. Single nucleotide variants (SNVs), insertions/deletions (indels), and copy number alterations (CNAs) were identified. We then evaluated the associations between each gene alteration with a frequency of 10% or more and progression-free survival (PFS) using a Cox regression model. We controlled for family-wise errors at 0.05 using the Bonferroni method. Among the 379 patients who were enrolled in JCOG0502, 127 patients were successfully analyzed using WES. The median patient age was 63 years (interquartile range, 57-67 years), and 78.0% of the patients ultimately underwent surgery. The 3-year PFS probability was 76.3%. We detected 20 genes with SNVs, indels, or amplifications with a frequency of 10% or more. Genomic alterations in FGF19 showed the strongest association with PFS with a borderline level of statistical significance of P = .00252 (Bonferroni-adjusted significance level is .0025). Genomic alterations in FGF4, MYEOV, CTTN, and ORAOV1 showed a marginal association with PFS (P < .05). These genomic alterations were all CNAs at chromosome 11q13.3. We have identified new genomic alterations associated with the poor efficacy of ESCC (T1bN0M0). These findings open avenues for the development of new potential treatments for patients with ESCC.
Keywords: esophageal squamous cell carcinoma; prognostic factor; stage I; tumor mutation burden; whole-exome sequencing.
© 2021 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Figures

References
-
- Engel LS, Chow WH, Vaughan TL, et al. Population attributable risks of esophageal and gastric cancers. J Natl Cancer Inst. 2003;95:1404‐1413. - PubMed
-
- Yokoyama T, Yokoyama A, Kato H, et al. Alcohol flushing, alcohol and aldehyde dehydrogenase genotypes, and risk for esophageal squamous cell carcinoma in Japanese men. Cancer Epidemiol Biomarkers Prev. 2003;12:1227‐1233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

