Photosynthesis-related genes induce resistance against soybean mosaic virus: Evidence for involvement of the RNA silencing pathway
- PMID: 34962034
- PMCID: PMC8916206
- DOI: 10.1111/mpp.13177
Photosynthesis-related genes induce resistance against soybean mosaic virus: Evidence for involvement of the RNA silencing pathway
Abstract
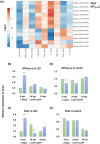
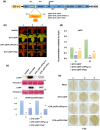
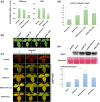
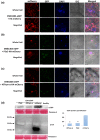
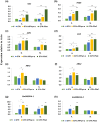
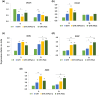
Increasing lines of evidence indicate that chloroplast-related genes are involved in plant-virus interactions. However, the involvement of photosynthesis-related genes in plant immunity is largely unexplored. Analysis of RNA-Seq data from the soybean cultivar L29, which carries the Rsv3 resistance gene, showed that several chloroplast-related genes were strongly induced in response to infection with an avirulent strain of soybean mosaic virus (SMV), G5H, but were weakly induced in response to a virulent strain, G7H. For further analysis, we selected the PSaC gene from the photosystem I and the ATP-synthase α-subunit (ATPsyn-α) gene whose encoded protein is part of the ATP-synthase complex. Overexpression of either gene within the G7H genome reduced virus levels in the susceptible cultivar Lee74 (rsv3-null). This result was confirmed by transiently expressing both genes in Nicotiana benthamiana followed by G7H infection. Both proteins localized in the chloroplast envelope as well as in the nucleus and cytoplasm. Because the chloroplast is the initial biosynthesis site of defence-related hormones, we determined whether hormone-related genes are involved in the ATPsyn-α- and PSaC-mediated defence. Interestingly, genes involved in the biosynthesis of several hormones were up-regulated in plants infected with SMV-G7H expressing ATPsyn-α. However, only jasmonic and salicylic acid biosynthesis genes were up-regulated following infection with the SMV-G7H expressing PSaC. Both chimeras induced the expression of several antiviral RNA silencing genes, which indicate that such resistance may be partially achieved through the RNA silencing pathway. These findings highlight the role of photosynthesis-related genes in regulating resistance to viruses.
Keywords: ATPsyn-α; PSaC; RNA silencing; photosynthesis; plant hormones; plant-virus interactions; soybean; soybean mosaic virus.
© 2021 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Chloroplast-related host proteins interact with NIb and NIa-Pro of soybeans mosaic virus and induce resistance in the susceptible cultivar.Virus Res. 2023 Oct 15;336:199205. doi: 10.1016/j.virusres.2023.199205. Epub 2023 Aug 25. Virus Res. 2023. PMID: 37607595 Free PMC article.
-
Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus.Viruses. 2018 Oct 24;10(11):581. doi: 10.3390/v10110581. Viruses. 2018. PMID: 30355968 Free PMC article.
-
An Avirulent Strain of Soybean Mosaic Virus Reverses the Defensive Effect of Abscisic Acid in a Susceptible Soybean Cultivar.Viruses. 2019 Sep 19;11(9):879. doi: 10.3390/v11090879. Viruses. 2019. PMID: 31546878 Free PMC article.
-
Decades of Genetic Research on Soybean mosaic virus Resistance in Soybean.Viruses. 2022 May 24;14(6):1122. doi: 10.3390/v14061122. Viruses. 2022. PMID: 35746594 Free PMC article. Review.
-
The Crucial Role of Chloroplast-Related Proteins in Viral Genome Replication and Host Defense against Positive-Sense Single-Stranded RNA Viruses.Plant Pathol J. 2023 Feb;39(1):28-38. doi: 10.5423/PPJ.RW.10.2022.0139. Epub 2023 Feb 1. Plant Pathol J. 2023. PMID: 36760047 Free PMC article. Review.
Cited by
-
Plant protection from virus: a review of different approaches.Front Plant Sci. 2023 Jun 12;14:1163270. doi: 10.3389/fpls.2023.1163270. eCollection 2023. Front Plant Sci. 2023. PMID: 37377807 Free PMC article. Review.
-
Transcriptomic profiling reveals the complex interaction between a bipartite begomovirus and a cucurbitaceous host plant.BMC Genomics. 2024 Sep 18;25(1):876. doi: 10.1186/s12864-024-10781-6. BMC Genomics. 2024. PMID: 39294575 Free PMC article.
-
A 4D Proteome Investigation of the Potential Mechanisms of SA in Triggering Resistance in Kiwifruit to Pseudomonas syringae pv. actinidiae.Int J Mol Sci. 2023 Dec 13;24(24):17448. doi: 10.3390/ijms242417448. Int J Mol Sci. 2023. PMID: 38139278 Free PMC article.
-
Resistance-breaking strains of tomato spotted wilt virus hamper photosynthesis and protein synthesis pathways in a virus accumulation-dependent manner in Sw5-carrying tomatoes.Sci Rep. 2025 Jan 29;15(1):3630. doi: 10.1038/s41598-025-88028-x. Sci Rep. 2025. PMID: 39881192 Free PMC article.
-
Novel 1,3,4-Thiadiazole Derivatives: Synthesis, Antiviral Bioassay and Regulation the Photosynthetic Pathway of Tobacco against TMV Infection.Int J Mol Sci. 2023 May 17;24(10):8881. doi: 10.3390/ijms24108881. Int J Mol Sci. 2023. PMID: 37240228 Free PMC article.
References
-
- Alazem, M. & Lin, N.S. (2020) Interplay between ABA signaling and RNA silencing in plant viral resistance. Current Opinion in Virology, 42, 1–7. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

