Spatial-temporal distribution and sequence diversity of group a human respiratory syncytial viruses in Kenya preceding the emergence of ON1 genotype
- PMID: 34962085
- PMCID: PMC8983921
- DOI: 10.1111/irv.12948
Spatial-temporal distribution and sequence diversity of group a human respiratory syncytial viruses in Kenya preceding the emergence of ON1 genotype
Abstract
Background: Human respiratory syncytial virus (HRSV) is a major cause of severe viral acute respiratory illness and contributes significantly to severe pneumonia cases in Africa. Little is known about its spatial-temporal distribution as defined by its genetic diversity.
Methods: A retrospective study conducted utilizing archived nasopharyngeal specimens from patients attending outpatient clinics in hospitals located in five demographically and climatically distinct regions of Kenya; Coast, Western, Highlands, Eastern and Nairobi. The viral total RNA was extracted and tested using multiplex real time RT-PCR (reverse transcriptase polymerase chain reaction). A segment of the G-gene was amplified using one-step RT-PCR and sequenced by Sanger di-deoxy method. Bayesian analysis of phylogeny was utilized and subsequently median joining methods for haplotype network reconstruction.
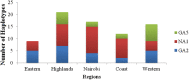
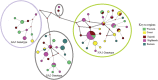
Results: Three genotypes of HRSVA were detected; GA5 (14.0%), GA2 (33.1%), and NA1 (52.9%). HRSVA prevalence varied by location from 33% to 13.2% in the Highlands and the Eastern regions respectively. The mean nucleotide diversity (Pi[π]) varied by genotype: highest of 0.018 for GA5 and lowest of 0.005 for NA1. A total of 58 haplotypes were identified (GA5 10; GA2 20; NA1 28). These haplotypes were introduced into the population locally by single haplotypes and additional subsidiary seeds amongst the GA2 and the NA1 haplotypes.
Conclusions: HRSVA was found across all the regions throughout the study period and comprised three genotypes; GA5, GA2, and NA1 genotypes. The genotypes were disproportionately distributed across the regions with GA5 gradually increasing toward the Western zones and decreasing toward the Eastern zones of the country.
Keywords: HRSVA; genotypes; haplotypes; regions.
© 2021 The Authors. Influenza and Other Respiratory Viruses published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Cane PA. Molecular Epidemiology of Respiratory Syncytial Virus. Rev Med Virol. 2001;11(2):103‐116. - PubMed
-
- Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ. Circulation patterns of genetically distinct group a and b strains of human respiratory syncytial virus in a community. J Gen Virol. 1998;79(Pt 9):2221‐2229. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

