Global Emergence and Dissemination of Neisseria gonorrhoeae ST-9363 Isolates with Reduced Susceptibility to Azithromycin
- PMID: 34962987
- PMCID: PMC8778598
- DOI: 10.1093/gbe/evab287
Global Emergence and Dissemination of Neisseria gonorrhoeae ST-9363 Isolates with Reduced Susceptibility to Azithromycin
Abstract
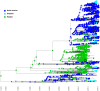
Neisseria gonorrhoeae multilocus sequence type (ST) 9363 core-genogroup isolates have been associated with reduced azithromycin susceptibility (AZMrs) and show evidence of clonal expansion in the United States. Here, we analyze a global collection of ST-9363 core-genogroup genomes to shed light on the emergence and dissemination of this strain. The global population structure of ST-9363 core-genogroup falls into three lineages: Basal, European, and North American; with 32 clades within all lineages. Although, ST-9363 core-genogroup is inferred to have originated from Asia in the mid-19th century; we estimate the three modern lineages emerged from Europe in the late 1970s to early 1980s. The European lineage appears to have emerged and expanded from around 1986 to 1998, spreading into North America and Oceania in the mid-2000s with multiple introductions, along with multiple secondary reintroductions into Europe. Our results suggest two separate acquisition events of mosaic mtrR and mtrR promoter alleles: first during 2009-2011 and again during the 2012-2013 time, facilitating the clonal expansion of this core-genogroup with AZMrs in the United States. By tracking phylodynamic evolutionary trajectories of clades that share distinct demography as well as population-based genomic statistics, we demonstrate how recombination and selective pressures in the mtrCDE efflux operon granted a fitness advantage to establish ST-9363 as a successful gonococcal lineage in the United States and elsewhere. Although it is difficult to pinpoint the exact timing and emergence of this young core-genogroup, it remains critically important to continue monitoring it, as it could acquire additional resistance markers.
Keywords: Neisseria gonorrhoeae; ST-9363; antibiotic resistance; azithromycin; evolution; phylogeography; recombination; timed phylogeny.
Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution 2021.
Figures



References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. - PubMed
-
- Banhart S, et al. 2021. The mosaic mtr locus as major genetic determinant of azithromycin resistance of Neisseria gonorrhoeae, Germany, 2018. J Infect Dis. 224(8):1398–1404. - PubMed
-
- Biswas S, Akey JM. 2006. Genomic insights into positive selection. Trends Genet. 22(8):437–446. - PubMed
-
- Bowen VB, et al. 2019. Sexually transmitted disease surveillance 2018. Available from: https://stacks.cdc.gov/view/cdc/79370. Accessed July 4, 2021.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

