Molecular detection of SARS-CoV-2 strains and differentiation of Delta variant strains
- PMID: 34964565
- PMCID: PMC9240106
- DOI: 10.1111/tbed.14443
Molecular detection of SARS-CoV-2 strains and differentiation of Delta variant strains
Abstract
The Delta variant of SARS-CoV-2 has now become the predominant strain in the global COVID-19 pandemic. Strain coverage of some detection assays developed during the early pandemic stages has declined due to periodic mutations in the viral genome. We have developed a real-time RT-PCR (RT-qPCR) for SARS-CoV-2 detection that provides nearly 100% strain coverage, and differentiation of highly transmissible Delta variant strains. All full or nearly full (≥28 kb) SARS-CoV-2 genomes (n = 403,812), including 6422 Delta and 280 Omicron variant strains, were collected from public databases at the time of analysis and used for assay design. The two amino acid deletions in the spike gene (S-gene, Δ156-157) that is characteristic of the Delta variant were targeted during the assay design. Although strain coverage for the Delta variant was very high (99.7%), detection coverage for non-Delta wild-type strains was 93.9%, mainly due to the confined region of design. To increase strain coverage of the assay, the design for CDC N1 target was added to the assay. In silico analysis of 403,812 genomes indicated a 95.4% strain coverage for the CDC N1 target, however, in combination with our new non-Delta S-gene target, total coverage for non-Delta wild-type strains increased to 99.8%. A human 18S rRNA gene was also analyzed and used as an internal control. The final four-plex RT-qPCR assay generated PCR amplification efficiencies between 95.4% and 102.0% with correlation coefficients (R2 ) of >0.99 for cloned positive controls; Delta and non-Delta human clinical samples generated PCR efficiencies of 93.4%-97.0% and R2 > 0.99. The assay also detects 98.6% of 280 Omicron sequences. Assay primers and probes have no match to other closely related human coronaviruses, and did not produce a signal from samples positive to selected animal coronaviruses. Genotypes of selected clinical samples identified by the RT-qPCR were confirmed by Sanger sequencing.
Keywords: COVID-19; Delta variant; Omicron variant; PCR; SARS-CoV-2; assay; diagnosis.
© 2022 Wiley-VCH GmbH.
Conflict of interest statement
Figures

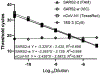

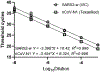

References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

