The polymorphism L412F in TLR3 inhibits autophagy and is a marker of severe COVID-19 in males
- PMID: 34964709
- PMCID: PMC9298458
- DOI: 10.1080/15548627.2021.1995152
The polymorphism L412F in TLR3 inhibits autophagy and is a marker of severe COVID-19 in males
Abstract
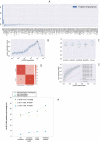
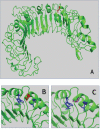
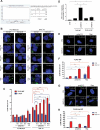
The polymorphism L412F in TLR3 has been associated with several infectious diseases. However, the mechanism underlying this association is still unexplored. Here, we show that the L412F polymorphism in TLR3 is a marker of severity in COVID-19. This association increases in the sub-cohort of males. Impaired macroautophagy/autophagy and reduced TNF/TNFα production was demonstrated in HEK293 cells transfected with TLR3L412F-encoding plasmid and stimulated with specific agonist poly(I:C). A statistically significant reduced survival at 28 days was shown in L412F COVID-19 patients treated with the autophagy-inhibitor hydroxychloroquine (p = 0.038). An increased frequency of autoimmune disorders such as co-morbidity was found in L412F COVID-19 males with specific class II HLA haplotypes prone to autoantigen presentation. Our analyses indicate that L412F polymorphism makes males at risk of severe COVID-19 and provides a rationale for reinterpreting clinical trials considering autophagy pathways.Abbreviations: AP: autophagosome; AUC: area under the curve; BafA1: bafilomycin A1; COVID-19: coronavirus disease-2019; HCQ: hydroxychloroquine; RAP: rapamycin; ROC: receiver operating characteristic; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; TLR: toll like receptor; TNF/TNF-α: tumor necrosis factor.
Keywords: Autophagy; COVID-19; HLA; L412F; TLR3.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
References
-
- Mukherjee S, Huda S, Sinha Babu SP.. Toll-like receptor polymorphism in host immune response to infectious diseases: a review. Scand J Immunol. 2019. Jul;90(1):e12771. - PubMed
-
- Matsumoto M, Oshiumi H, Seya T. Antiviral responses induced by the TLR3 pathway. Rev Med Virol. 2011. Mar;21(2):67–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
