AMPK-mTOR-Mediated Activation of Autophagy Promotes Formation of Dormant Polyploid Giant Cancer Cells
- PMID: 34965934
- PMCID: PMC9359740
- DOI: 10.1158/0008-5472.CAN-21-2342
AMPK-mTOR-Mediated Activation of Autophagy Promotes Formation of Dormant Polyploid Giant Cancer Cells
Abstract
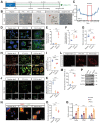
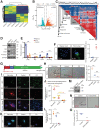
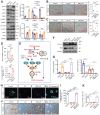
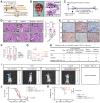
Dormant cancer cells that survive anticancer therapy can lead to cancer recurrence and disseminated metastases that prove fatal in most cases. Recently, specific dormant polyploid giant cancer cells (PGCC) have drawn our attention because of their association with the clinical risk of nasopharyngeal carcinoma (NPC) recurrence, as demonstrated by previous clinical data. In this study, we report the biological properties of PGCC, including mitochondrial alterations, and reveal that autophagy is a critical mechanism of PGCC induction. Moreover, pharmacologic or genetic inhibition of autophagy greatly impaired PGCC formation, significantly suppressing metastasis and improving survival in a mouse model. Mechanistically, chemotherapeutic drugs partly damaged mitochondria, which then produced low ATP levels and activated autophagy via the AMPK-mTOR pathway to promote PGCC formation. Analysis of the transcriptional and epigenetic landscape of PGCC revealed overexpression of RIPK1, and the scaffolding function of RIPK1 was required for AMPK-mTOR pathway-induced PGCC survival. High numbers of PGCCs correlated with shorter recurrence time and worse survival outcomes in patients with NPC. Collectively, these findings suggest a therapeutic approach of targeting dormant PGCCs in cancer.
Significance: Pretreatment with an autophagy inhibitor before chemotherapy could prevent formation of therapy-induced dormant polyploid giant cancer cells, thereby reducing recurrence and metastasis of nasopharyngeal carcinoma.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures

References
-
- Zhang L, Huang Y, Hong S, Yang Y, Yu G, Jia J, et al. . Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: a multicentre, randomised, open-label, phase 3 trial. Lancet 2016;388:1883–92. - PubMed
-
- Phan TG, Croucher PI. The dormant cancer cell life cycle. Nat Rev Cancer 2020;20:398–411. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

