A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants
- PMID: 34966387
- PMCID: PMC8710476
- DOI: 10.3389/fimmu.2021.766821
A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants
Abstract
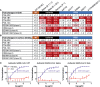
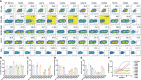
As severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants continue to emerge and spread around the world, antibodies and vaccines to confer broad and potent neutralizing activity are urgently needed. Through the isolation and characterization of monoclonal antibodies (mAbs) from individuals infected with SARS-CoV-2, we identified one antibody, P36-5D2, capable of neutralizing the major SARS-CoV-2 variants of concern. Crystal and electron cryo-microscopy (cryo-EM) structure analyses revealed that P36-5D2 targeted to a conserved epitope on the receptor-binding domain of the spike protein, withstanding the three key mutations-K417N, E484K, and N501Y-found in the variants that are responsible for escape from many potent neutralizing mAbs, including some already approved for emergency use authorization (EUA). A single intraperitoneal (IP) injection of P36-5D2 as a prophylactic treatment completely protected animals from challenge of infectious SARS-CoV-2 Alpha and Beta. Treated animals manifested normal body weight and were devoid of infection-associated death up to 14 days. A substantial decrease of the infectious virus in the lungs and brain, as well as reduced lung pathology, was found in these animals compared to the controls. Thus, P36-5D2 represents a new and desirable human antibody against the current and emerging SARS-CoV-2 variants.
Keywords: SARS-CoV-2; epitope; human neutralizing antibody; in vivo protection; variants of concern.
Copyright © 2021 Shan, Mok, Zhang, Lan, Li, Yang, Wang, Cheng, Fang, Aw, Yu, Zhang, Shi, Zhang, Zhang, Wang, Wang, Chu and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Characterization of MW06, a human monoclonal antibody with cross-neutralization activity against both SARS-CoV-2 and SARS-CoV.MAbs. 2021 Jan-Dec;13(1):1953683. doi: 10.1080/19420862.2021.1953683. MAbs. 2021. PMID: 34313527 Free PMC article.
-
Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.Nature. 2020 Jul;583(7815):290-295. doi: 10.1038/s41586-020-2349-y. Epub 2020 May 18. Nature. 2020. PMID: 32422645
-
Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.Nat Commun. 2021 Jan 11;12(1):264. doi: 10.1038/s41467-020-20465-w. Nat Commun. 2021. PMID: 33431876 Free PMC article.
-
Passive antibody therapy in emerging infectious diseases.Front Med. 2023 Dec;17(6):1117-1134. doi: 10.1007/s11684-023-1021-y. Epub 2023 Dec 2. Front Med. 2023. PMID: 38040914 Review.
-
Effective high-throughput isolation of fully human antibodies targeting infectious pathogens.Nat Protoc. 2021 Jul;16(7):3639-3671. doi: 10.1038/s41596-021-00554-w. Epub 2021 May 25. Nat Protoc. 2021. PMID: 34035500 Review.
Cited by
-
Super broad and protective nanobodies against Sarbecoviruses including SARS-CoV-1 and the divergent SARS-CoV-2 subvariant KP.3.1.1.PLoS Pathog. 2024 Nov 11;20(11):e1012625. doi: 10.1371/journal.ppat.1012625. eCollection 2024 Nov. PLoS Pathog. 2024. PMID: 39527594 Free PMC article.
-
Novel bispecific human antibody platform specifically targeting a fully open spike conformation potently neutralizes multiple SARS-CoV-2 variants.Antiviral Res. 2023 Apr;212:105576. doi: 10.1016/j.antiviral.2023.105576. Epub 2023 Mar 2. Antiviral Res. 2023. PMID: 36870394 Free PMC article.
-
Deep learning guided optimization of human antibody against SARS-CoV-2 variants with broad neutralization.Proc Natl Acad Sci U S A. 2022 Mar 15;119(11):e2122954119. doi: 10.1073/pnas.2122954119. Epub 2022 Mar 1. Proc Natl Acad Sci U S A. 2022. PMID: 35238654 Free PMC article.
-
The humoral response and antibodies against SARS-CoV-2 infection.Nat Immunol. 2022 Jul;23(7):1008-1020. doi: 10.1038/s41590-022-01248-5. Epub 2022 Jun 27. Nat Immunol. 2022. PMID: 35761083 Review.
-
Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.Cell Discov. 2022 Sep 8;8(1):87. doi: 10.1038/s41421-022-00449-4. Cell Discov. 2022. PMID: 36075908 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

