Oligosarcomas, IDH-mutant are distinct and aggressive
- PMID: 34967922
- PMCID: PMC8742817
- DOI: 10.1007/s00401-021-02395-z
Oligosarcomas, IDH-mutant are distinct and aggressive
Abstract
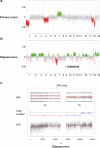
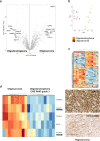
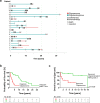
Oligodendrogliomas are defined at the molecular level by the presence of an IDH mutation and codeletion of chromosomal arms 1p and 19q. In the past, case reports and small studies described gliomas with sarcomatous features arising from oligodendrogliomas, so called oligosarcomas. Here, we report a series of 24 IDH-mutant oligosarcomas from 23 patients forming a distinct methylation class. The tumors were recurrences from prior oligodendrogliomas or developed de novo. Precursor tumors of 12 oligosarcomas were histologically and molecularly indistinguishable from conventional oligodendrogliomas. Oligosarcoma tumor cells were embedded in a dense network of reticulin fibers, frequently showing p53 accumulation, positivity for SMA and CALD1, loss of OLIG2 and gain of H3K27 trimethylation (H3K27me3) as compared to primary lesions. In 5 oligosarcomas no 1p/19q codeletion was detectable, although it was present in the primary lesions. Copy number neutral LOH was determined as underlying mechanism. Oligosarcomas harbored an increased chromosomal copy number variation load with frequent CDKN2A/B deletions. Proteomic profiling demonstrated oligosarcomas to be highly distinct from conventional CNS WHO grade 3 oligodendrogliomas with consistent evidence for a smooth muscle differentiation. Expression of several tumor suppressors was reduced with NF1 being lost frequently. In contrast, oncogenic YAP1 was aberrantly overexpressed in oligosarcomas. Panel sequencing revealed mutations in NF1 and TP53 along with IDH1/2 and TERT promoter mutations. Survival of patients was significantly poorer for oligosarcomas as first recurrence than for grade 3 oligodendrogliomas as first recurrence. These results establish oligosarcomas as a distinct group of IDH-mutant gliomas differing from conventional oligodendrogliomas on the histologic, epigenetic, proteomic, molecular and clinical level. The diagnosis can be based on the combined presence of (a) sarcomatous histology, (b) IDH-mutation and (c) TERT promoter mutation and/or 1p/19q codeletion, or, in unresolved cases, on its characteristic DNA methylation profile.
Keywords: 1p/19q; Codeletion; DNA methylation; Gliosarcoma; NF1; Oligodendroglioma; Oligosarcoma; Prognosis; SMA; Subtype; TERT; TP53; Type; Variant; YAP1.
© 2021. The Author(s).
Conflict of interest statement
Under a licensing agreement between DIANOVA GmbH, Hamburg, Germany, and the German Cancer Research Center, DC, CH, AVD are entitled to a share of royalties received by the German Cancer Research Center on the sales of H09 antibody. The terms of this arrangement are being managed by the German Cancer Research Center in accordance with its conflict of interest policies. AVD and DER have licensed the NFC antibody to Merck Millipore. All terms are being managed by the University of Heidelberg in accordance with its conflict of interest policies.
Figures




References
-
- Agozzino L, Pittore L, Lamendola MG, Ambrosio A. Sarcoma arising in a cerebral oligodendroglioma. Case report. Pathologica. 1983;75:501–508. - PubMed
-
- Bady P, Sciuscio D, Diserens AC, Bloch J, van den Bent MJ, Marosi C, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124:547–560. doi: 10.1007/s00401-012-1016-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

