H3K4 Methylation in Aging and Metabolism
- PMID: 34968301
- PMCID: PMC8594702
- DOI: 10.3390/epigenomes5020014
H3K4 Methylation in Aging and Metabolism
Abstract
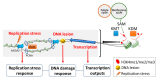
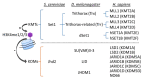
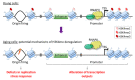
During the process of aging, extensive epigenetic alterations are made in response to both exogenous and endogenous stimuli. Here, we summarize the current state of knowledge regarding one such alteration, H3K4 methylation (H3K4me), as it relates to aging in different species. We especially highlight emerging evidence that links this modification with metabolic pathways, which may provide a mechanistic link to explain its role in aging. H3K4me is a widely recognized marker of active transcription, and it appears to play an evolutionarily conserved role in determining organism longevity, though its influence is context specific and requires further clarification. Interestingly, the modulation of H3K4me dynamics may occur as a result of nutritional status, such as methionine restriction. Methionine status appears to influence H3K4me via changes in the level of S-adenosyl methionine (SAM, the universal methyl donor) or the regulation of H3K4-modifying enzyme activities. Since methionine restriction is widely known to extend lifespan, the mechanistic link between methionine metabolic flux, the sensing of methionine concentrations and H3K4me status may provide a cogent explanation for several seemingly disparate observations in aging organisms, including age-dependent H3K4me dynamics, gene expression changes, and physiological aberrations. These connections are not yet entirely understood, especially at a molecular level, and will require further elucidation. To conclude, we discuss some potential H3K4me-mediated molecular mechanisms that may link metabolic status to the aging process.
Keywords: H3K4 methylation; aging; metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

