Investigation of a SARS-CoV-2 B.1.1.529 (Omicron) Variant Cluster - Nebraska, November-December 2021
- PMID: 34968376
- PMCID: PMC8736273
- DOI: 10.15585/mmwr.mm705152e3
Investigation of a SARS-CoV-2 B.1.1.529 (Omicron) Variant Cluster - Nebraska, November-December 2021
Abstract
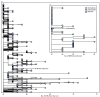
The B.1.1.529 (Omicron) variant of SARS-CoV-2 (the virus that causes COVID-19) was first detected in specimens collected on November 11, 2021, in Botswana and on November 14 in South Africa;* the first confirmed case of Omicron in the United States was identified in California on December 1, 2021 (1). On November 29, the Nebraska Department of Health and Human Services was notified of six probable cases† of COVID-19 in one household, including one case in a man aged 48 years (the index patient) who had recently returned from Nigeria. Given the patient's travel history, Omicron infection was suspected. Specimens from all six persons in the household tested positive for SARS-CoV-2 by reverse transcription-polymerase chain reaction (RT-PCR) testing on December 1, and the following day genomic sequencing by the Nebraska Public Health Laboratory identified an identical Omicron genotype from each specimen (Figure). Phylogenetic analysis was conducted to determine if this cluster represented an independent introduction of Omicron into the United States, and a detailed epidemiologic investigation was conducted. This activity was reviewed by CDC and was conducted consistent with applicable federal law and CDC policy.§.
Conflict of interest statement
All authors have completed and submitted the International Committee of Medical Journal Editors form for disclosure of potential conflicts of interest. Joseph Fauver reports that he is a consultant with Tempus Labs to conduct SARS-CoV-2 genomic surveillance in the National Football League. Peter C. Iwen reports support from the American Rescue Plan Act of 2021 ELC Epi and Lab Capacity for Genomic Sequencing from the U.S. Department of Health and Human Services and the Paycheck Protection Program and Health Care Enhancement Act ELC Epi and Lab Capacity from the U.S. Department of Health and Human Services. No other potential conflicts of interest were disclosed.
Figures
References
-
- Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (CoVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med 2020;172:577–82. PMID:32150748.10.7326/M20-0504 - DOI - PMC - PubMed
-
- Grant R, Charmet T, Schaeffer L, et al. Impact of SARS-CoV-2 Delta variant on incubation, transmission settings and vaccine effectiveness: results from a nationwide case-control study in France. Lancet Reg Heal Eur 2021. Epub November 25, 2021. https://www.thelancet.com/article/S2666776221002647/fulltext - PMC - PubMed
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

