A Recent Update on Advanced Molecular Diagnostic Techniques for COVID-19 Pandemic: An Overview
- PMID: 34970254
- PMCID: PMC8712736
- DOI: 10.3389/fimmu.2021.732756
A Recent Update on Advanced Molecular Diagnostic Techniques for COVID-19 Pandemic: An Overview
Abstract
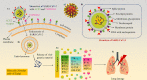
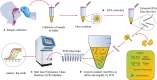
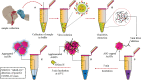
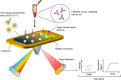
Coronavirus disease 2019 (COVID-19), which started out as an outbreak of pneumonia, has now turned into a pandemic due to its rapid transmission. Besides developing a vaccine, rapid, accurate, and cost-effective diagnosis is essential for monitoring and combating the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its related variants on time with precision and accuracy. Currently, the gold standard for detection of SARS-CoV-2 is Reverse Transcription Polymerase Chain Reaction (RT-PCR), but it lacks accuracy, is time-consuming and cumbersome, and fails to detect multi-variant forms of the virus. Herein, we have summarized conventional diagnostic methods such as Chest-CT (Computed Tomography), RT-PCR, Loop Mediated Isothermal Amplification (LAMP), Reverse Transcription-LAMP (RT-LAMP), as well new modern diagnostics such as CRISPR-Cas-based assays, Surface Enhanced Raman Spectroscopy (SERS), Lateral Flow Assays (LFA), Graphene-Field Effect Transistor (GraFET), electrochemical sensors, immunosensors, antisense oligonucleotides (ASOs)-based assays, and microarrays for SARS-CoV-2 detection. This review will also provide an insight into an ongoing research and the possibility of developing more economical tools to tackle the COVID-19 pandemic.
Keywords: CRISPR-Cas; LAMP; RT-PCR; SARS-CoV-2; biosensors; diagnostics; point of care.
Copyright © 2021 Roberts, Chouhan, Shahdeo, Shrikrishna, Kesarwani, Horvat and Gandhi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Development and Clinical Application of a Rapid and Sensitive Loop-Mediated Isothermal Amplification Test for SARS-CoV-2 Infection.mSphere. 2020 Aug 26;5(4):e00808-20. doi: 10.1128/mSphere.00808-20. mSphere. 2020. PMID: 32848011 Free PMC article.
-
A semi-automated, isolation-free, high-throughput SARS-CoV-2 reverse transcriptase (RT) loop-mediated isothermal amplification (LAMP) test.Sci Rep. 2021 Nov 1;11(1):21385. doi: 10.1038/s41598-021-00827-0. Sci Rep. 2021. PMID: 34725400 Free PMC article.
-
Rapid, Sensitive, and Specific Severe Acute Respiratory Syndrome Coronavirus 2 Detection: A Multicenter Comparison Between Standard Quantitative Reverse-Transcriptase Polymerase Chain Reaction and CRISPR-Based DETECTR.J Infect Dis. 2021 Feb 3;223(2):206-213. doi: 10.1093/infdis/jiaa641. J Infect Dis. 2021. PMID: 33535237 Free PMC article.
-
Diagnostic efficiency of RT-LAMP integrated CRISPR-Cas technique for COVID-19: A systematic review and meta-analysis.Pathog Glob Health. 2022 Oct;116(7):410-420. doi: 10.1080/20477724.2022.2035625. Epub 2022 Feb 10. Pathog Glob Health. 2022. PMID: 35142264 Free PMC article.
-
Advancements in detection of SARS-CoV-2 infection for confronting COVID-19 pandemics.Lab Invest. 2022 Jan;102(1):4-13. doi: 10.1038/s41374-021-00663-w. Epub 2021 Sep 8. Lab Invest. 2022. PMID: 34497366 Free PMC article. Review.
Cited by
-
Production of a Monoclonal Antibody to the Nucleocapsid Protein of SARS-CoV-2 and Its Application to ELISA-Based Detection Methods with Broad Specificity by Combined Use of Detector Antibodies.Viruses. 2022 Dec 21;15(1):28. doi: 10.3390/v15010028. Viruses. 2022. PMID: 36680068 Free PMC article.
-
SERS-Based Biosensors Combined with Machine Learning for Medical Application.ChemistryOpen. 2023 Jan;12(1):e202200192. doi: 10.1002/open.202200192. ChemistryOpen. 2023. PMID: 36627171 Free PMC article. Review.
-
Emerging 0D, 1D, 2D, and 3D nanostructures for efficient point-of-care biosensing.Biosens Bioelectron X. 2022 Dec;12:100284. doi: 10.1016/j.biosx.2022.100284. Epub 2022 Nov 25. Biosens Bioelectron X. 2022. PMID: 36448023 Free PMC article.
-
Performance Evaluation of the STANDARD i-Q COVID-19 Ag Test with Nasal and Oral Swab Specimens from Symptomatic Patients.Diagnostics (Basel). 2024 Jan 22;14(2):231. doi: 10.3390/diagnostics14020231. Diagnostics (Basel). 2024. PMID: 38275478 Free PMC article.
-
Nanomaterials-based sensors for the detection of COVID-19: A review.Bioeng Transl Med. 2022 Apr 13;7(3):e10305. doi: 10.1002/btm2.10305. eCollection 2022 Sep. Bioeng Transl Med. 2022. PMID: 35599642 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

