Multiplex-Heterogeneous Network-Based Capturing Potential SNP "Switches" of Pathways Associating With Diverse Disease Characteristics of Asthma
- PMID: 34970542
- PMCID: PMC8712737
- DOI: 10.3389/fcell.2021.744932
Multiplex-Heterogeneous Network-Based Capturing Potential SNP "Switches" of Pathways Associating With Diverse Disease Characteristics of Asthma
Abstract
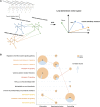
Asthma is a complex heterogeneous respiratory disorder. In recent years nubbly regions of the role of genetic variants and transcriptome including mRNAs, microRNAs, and long non-coding RNAs in the pathogenesis of asthma have been separately excavated and reported. However, how to systematically integrate and decode this scattered information remains unclear. Further exploration would improve understanding of the internal communication of asthma. To excavate new insights into the pathogenesis of asthma, we ascertained three asthma characteristics according to reviews, airway inflammation, airway hyperresponsiveness, and airway remodeling. We manually created a contemporary catalog of corresponding risk transcriptome, including mRNAs, miRNAs, and lncRNAs. MIMP is a multiplex-heterogeneous networks-based approach, measuring the relevance of disease characteristics to the pathway by examining the similarity between the determined vectors of risk transcriptome and pathways in the same low-dimensional vector space. It was developed to enable a more concentrated and in-depth exploration of potential pathways. We integrated experimentally validated competing endogenous RNA regulatory information and the SNPs with significant pathways into the ceRNA-mediated SNP switching pathway network (CSSPN) to analyze ceRNA regulation of pathways and the role of SNP in these dysfunctions. We discovered 11 crucial ceRNA regulations concerning asthma disease feature pathway and propose a potential mechanism of ceRNA regulatory SNP → gene → pathway → disease feature effecting asthma pathogenesis, especially for MALAT1 (rs765499057/rs764699354/rs189435941) → hsa-miR-155 → IL13 (rs201185816/rs1000978586/rs202101165) → Interleukin-4 and Interleukin-13 signaling → inflammation/airway remodeling and MALAT1 (rs765499057/rs764699354/rs189435941) → hsa-miR-155 → IL17RB (rs948046241) → Interleukin-17 signaling (airway remodeling)/Cytokine-cytokine receptor interaction (inflammation). This study showed a systematic and propagable workflow for capturing the potential SNP "switch" of asthma through text and database mining and provides further information on the pathogenesis of asthma.
Keywords: SNPs; asthma; bioinformatics; ceRNA; lncRNA; miRNA; network; pathway enrichment analysis.
Copyright © 2021 Ran, Yuan, Fan, Ke, Wang, Sun and Su.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Austin P. J., Tsitsiou E., Boardman C., Jones S. W., Lindsay M. A., Adcock I. M., et al. (2017). Transcriptional Profiling Identifies the Long Noncoding RNA Plasmacytoma Variant Translocation ( PVT1 ) as a Novel Regulator of the Asthmatic Phenotype in Human Airway Smooth Muscle. J. Allergy Clin. Immunol. 139, 780–789. 10.1016/j.jaci.2016.06.014 - DOI - PMC - PubMed
-
- Camargo L. d. N., Santos T. M. d., Andrade F. C. P. d., Fukuzaki S., Dos Santos Lopes F. D. T. Q., de Arruda Martins M., et al. (2020). Bronchial Vascular Remodeling Is Attenuated by Anti-IL-17 in Asthmatic Responses Exacerbated by LPS. Front. Pharmacol. 11, 1269. 10.3389/fphar.2020.01269 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

