The Mouse Gastrointestinal Bacteria Catalogue enables translation between the mouse and human gut microbiotas via functional mapping
- PMID: 34971560
- PMCID: PMC8763404
- DOI: 10.1016/j.chom.2021.12.003
The Mouse Gastrointestinal Bacteria Catalogue enables translation between the mouse and human gut microbiotas via functional mapping
Abstract
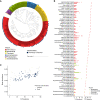
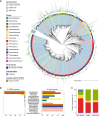
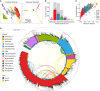
Human health and disease have increasingly been shown to be impacted by the gut microbiota, and mouse models are essential for investigating these effects. However, the compositions of human and mouse gut microbiotas are distinct, limiting translation of microbiota research between these hosts. To address this, we constructed the Mouse Gastrointestinal Bacteria Catalogue (MGBC), a repository of 26,640 high-quality mouse microbiota-derived bacterial genomes. This catalog enables species-level analyses for mapping functions of interest and identifying functionally equivalent taxa between the microbiotas of humans and mice. We have complemented this with a publicly deposited collection of 223 bacterial isolates, including 62 previously uncultured species, to facilitate experimental investigation of individual commensal bacteria functions in vitro and in vivo. Together, these resources provide the ability to identify and test functionally equivalent members of the host-specific gut microbiotas of humans and mice and support the informed use of mouse models in human microbiota research.
Keywords: bacteria culture collection; butyrate; commensal bacteria; functionally equivalent species; gut microbiota; microbial drug metabolism; mouse gut metagenomes; mouse models; public database; translation between mouse and human.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.D.L. is a founder and CSO of Microbiotica. The other authors declare no competinginterests.
Figures


References
-
- Aitchison J. On criteria for measures of compositional difference. Math. Geol. 1992;24:365–379. doi: 10.1007/BF00891269. - DOI
-
- Aluthge N.D., Tom W.A., Bartenslager A.C., Burkey T.E., Miller P.S., Heath K.D., Kreikemeier-Bower C., Kittana H., Schmaltz R.J., Ramer-Tait A.E., Fernando S.C. Differential longitudinal establishment of human fecal bacterial communities in germ-free porcine and murine models. Commun. Biol. 2020;3:760. doi: 10.1038/s42003-020-01477-0. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

