Hidden intermediates in Mango III RNA aptamer folding revealed by pressure perturbation
- PMID: 34971617
- PMCID: PMC8822612
- DOI: 10.1016/j.bpj.2021.12.037
Hidden intermediates in Mango III RNA aptamer folding revealed by pressure perturbation
Abstract
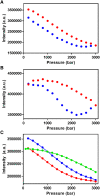
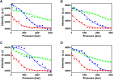
Fluorescent RNA aptamers have the potential to enable routine quantitation and localization of RNA molecules and serve as models for understanding biologically active aptamers. In recent years, several fluorescent aptamers have been selected and modified to improve their properties, revealing that small changes to the RNA or the ligands can modify significantly their fluorescent properties. Although structural biology approaches have revealed the bound, ground state of several fluorescent aptamers, characterization of low-abundance, excited states in these systems is crucial to understanding their folding pathways. Here we use pressure as an alternative variable to probe the suboptimal states of the Mango III aptamer with both fluorescence and NMR spectroscopy approaches. At moderate KCl concentrations, increasing pressure disrupted the G-quadruplex structure of the Mango III RNA and led to an intermediate with lower fluorescence. These observations indicate the existence of suboptimal RNA structural states that still bind the TO1-biotin fluorophore and moderately enhance fluorescence. At higher KCl concentration as well, the intermediate fluorescence state was populated at high pressure, but the G-quadruplex remained stable at high pressure, supporting the notion of parallel folding and/or binding pathways. These results demonstrate the usefulness of pressure for characterizing RNA folding intermediates.
Copyright © 2022 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Double-stemmed and split structural variants of fluorescent RNA Mango aptamers.RNA. 2023 Sep;29(9):1355-1364. doi: 10.1261/rna.079651.123. Epub 2023 Jun 2. RNA. 2023. PMID: 37268327 Free PMC article.
-
Turn-on RNA Mango Beacons for trans-acting fluorogenic nucleic acid detection.RNA. 2024 Mar 18;30(4):392-403. doi: 10.1261/rna.079833.123. RNA. 2024. PMID: 38282417 Free PMC article.
-
Fluorophore ligand binding and complex stabilization of the RNA Mango and RNA Spinach aptamers.RNA. 2016 Dec;22(12):1884-1892. doi: 10.1261/rna.056226.116. Epub 2016 Oct 24. RNA. 2016. PMID: 27777365 Free PMC article.
-
Tracking RNA with light: selection, structure, and design of fluorescence turn-on RNA aptamers.Q Rev Biophys. 2019 Aug 19;52:e8. doi: 10.1017/S0033583519000064. Q Rev Biophys. 2019. PMID: 31423956 Free PMC article. Review.
-
In Vivo Production of RNA Aptamers and Nanoparticles: Problems and Prospects.Molecules. 2021 Mar 6;26(5):1422. doi: 10.3390/molecules26051422. Molecules. 2021. PMID: 33800717 Free PMC article. Review.
Cited by
-
Pressure pushes tRNALys3 into excited conformational states.Proc Natl Acad Sci U S A. 2023 Jun 27;120(26):e2215556120. doi: 10.1073/pnas.2215556120. Epub 2023 Jun 20. Proc Natl Acad Sci U S A. 2023. PMID: 37339210 Free PMC article.
-
Double-stemmed and split structural variants of fluorescent RNA Mango aptamers.RNA. 2023 Sep;29(9):1355-1364. doi: 10.1261/rna.079651.123. Epub 2023 Jun 2. RNA. 2023. PMID: 37268327 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

