Improving local prevalence estimates of SARS-CoV-2 infections using a causal debiasing framework
- PMID: 34972825
- PMCID: PMC8727294
- DOI: 10.1038/s41564-021-01029-0
Improving local prevalence estimates of SARS-CoV-2 infections using a causal debiasing framework
Abstract
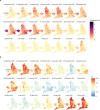
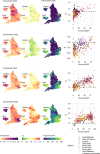
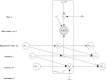
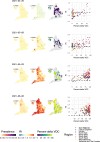
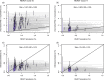
Global and national surveillance of SARS-CoV-2 epidemiology is mostly based on targeted schemes focused on testing individuals with symptoms. These tested groups are often unrepresentative of the wider population and exhibit test positivity rates that are biased upwards compared with the true population prevalence. Such data are routinely used to infer infection prevalence and the effective reproduction number, Rt, which affects public health policy. Here, we describe a causal framework that provides debiased fine-scale spatiotemporal estimates by combining targeted test counts with data from a randomized surveillance study in the United Kingdom called REACT. Our probabilistic model includes a bias parameter that captures the increased probability of an infected individual being tested, relative to a non-infected individual, and transforms observed test counts to debiased estimates of the true underlying local prevalence and Rt. We validated our approach on held-out REACT data over a 7-month period. Furthermore, our local estimates of Rt are indicative of 1-week- and 2-week-ahead changes in SARS-CoV-2-positive case numbers. We also observed increases in estimated local prevalence and Rt that reflect the spread of the Alpha and Delta variants. Our results illustrate how randomized surveys can augment targeted testing to improve statistical accuracy in monitoring the spread of emerging and ongoing infectious disease.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

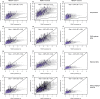
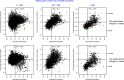
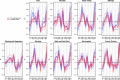
References
-
- PHE Data Series on Deaths in People with COVID-19: Technical Summary—12 August Update (Public Health England, 2020).
-
- The Official UK Government Website for Data and Insights on Coronavirus (COVID-19) (GOV.UK, accessed 15 February 2021); https://coronavirus.data.gov.uk
-
- Summary of Effectiveness and Harms of NPIs. Scientific Advisory Group for Emergencies (21 September 2020); https://www.gov.uk/government/publications/ summary-of-the-effectiveness...
-
- Prime Minister Announces New local COVID Alert Levels. Prime Minister’s Office, 10 Downing Street (12 October 2020); https://www.gov.uk/government/news/ prime-minister-announces-new-local- ...
-
- COVID-19 Response—Spring 2021 (Summary). Cabinet Office (22 February 2021); https://www.gov.uk/government/ publications/covid-19-response-spring-202...
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

