GranatumX: A Community-engaging, Modularized, and Flexible Webtool for Single-cell Data Analysis
- PMID: 34973417
- PMCID: PMC8864242
- DOI: 10.1016/j.gpb.2021.07.005
GranatumX: A Community-engaging, Modularized, and Flexible Webtool for Single-cell Data Analysis
Abstract
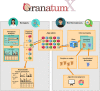
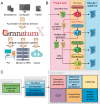
We present GranatumX, a next-generation software environment for single-cell RNA sequencing (scRNA-seq) data analysis. GranatumX is inspired by the interactive webtool Granatum. GranatumX enables biologists to access the latest scRNA-seq bioinformatics methods in a web-based graphical environment. It also offers software developers the opportunity to rapidly promote their own tools with others in customizable pipelines. The architecture of GranatumX allows for easy inclusion of plugin modules, named Gboxes, which wrap around bioinformatics tools written in various programming languages and on various platforms. GranatumX can be run on the cloud or private servers and generate reproducible results. It is a community-engaging, flexible, and evolving software ecosystem for scRNA-seq analysis, connecting developers with bench scientists. GranatumX is freely accessible at http://garmiregroup.org/granatumx/app.
Keywords: Analysis; Module; Pipeline; Single-cell RNA sequencing; Webtool.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Galaxy as a gateway to bioinformatics: Multi-Interface Galaxy Hands-on Training Suite (MIGHTS) for scRNA-seq.Gigascience. 2025 Jan 6;14:giae107. doi: 10.1093/gigascience/giae107. Gigascience. 2025. PMID: 39775842 Free PMC article.
-
Granatum: a graphical single-cell RNA-Seq analysis pipeline for genomics scientists.Genome Med. 2017 Dec 5;9(1):108. doi: 10.1186/s13073-017-0492-3. Genome Med. 2017. PMID: 29202807 Free PMC article.
-
Shaoxia: a web-based interactive analysis platform for single cell RNA sequencing data.BMC Genomics. 2024 Apr 24;25(1):402. doi: 10.1186/s12864-024-10322-1. BMC Genomics. 2024. PMID: 38658838 Free PMC article.
-
A guide to single-cell RNA sequencing analysis using web-based tools for non-bioinformatician.FEBS J. 2024 Jun;291(12):2545-2561. doi: 10.1111/febs.17036. Epub 2024 Jan 20. FEBS J. 2024. PMID: 38148322 Review.
-
Computational approaches for interpreting scRNA-seq data.FEBS Lett. 2017 Aug;591(15):2213-2225. doi: 10.1002/1873-3468.12684. Epub 2017 Jun 12. FEBS Lett. 2017. PMID: 28524227 Free PMC article. Review.
Cited by
-
Adding Highly Variable Genes to Spatially Variable Genes Can Improve Cell Type Clustering Performance in Spatial Transcriptomics Data.Res Sq [Preprint]. 2024 Oct 25:rs.3.rs-5315913. doi: 10.21203/rs.3.rs-5315913/v1. Res Sq. 2024. PMID: 39502778 Free PMC article. Preprint.
-
GRACE: a comprehensive web-based platform for integrative single-cell transcriptome analysis.NAR Genom Bioinform. 2023 Jun 9;5(2):lqad050. doi: 10.1093/nargab/lqad050. eCollection 2023 Jun. NAR Genom Bioinform. 2023. PMID: 37305171 Free PMC article.
-
Galaxy as a gateway to bioinformatics: Multi-Interface Galaxy Hands-on Training Suite (MIGHTS) for scRNA-seq.Gigascience. 2025 Jan 6;14:giae107. doi: 10.1093/gigascience/giae107. Gigascience. 2025. PMID: 39775842 Free PMC article.
-
Towards early diagnosis of Alzheimer's disease: advances in immune-related blood biomarkers and computational approaches.Front Immunol. 2024 Apr 23;15:1343900. doi: 10.3389/fimmu.2024.1343900. eCollection 2024. Front Immunol. 2024. PMID: 38720902 Free PMC article. Review.
-
Evaluation of Cell Type Annotation R Packages on Single-cell RNA-seq Data.Genomics Proteomics Bioinformatics. 2021 Apr;19(2):267-281. doi: 10.1016/j.gpb.2020.07.004. Epub 2020 Dec 24. Genomics Proteomics Bioinformatics. 2021. PMID: 33359678 Free PMC article.
References
-
- Svensson V., Vento-Tormo R., Teichmann S.A. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc. 2018;13(4):599–604. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

