Circular RNA protein tyrosine kinase 2 (circPTK2) promotes colorectal cancer proliferation, migration, invasion and chemoresistance
- PMID: 34974791
- PMCID: PMC8805883
- DOI: 10.1080/21655979.2021.2012952
Circular RNA protein tyrosine kinase 2 (circPTK2) promotes colorectal cancer proliferation, migration, invasion and chemoresistance
Retraction in
-
Statement of Retraction: Circular RNA protein tyrosine kinase 2 (circPTK2) promotes colorectal cancer proliferation, migration, invasion and chemoresistance.Bioengineered. 2023 Dec;14(1):2269352. doi: 10.1080/21655979.2023.2269352. Epub 2023 Oct 18. Bioengineered. 2023. PMID: 37853773 Free PMC article. No abstract available.
Abstract
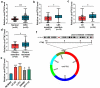
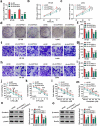
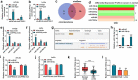
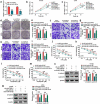
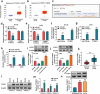
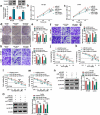
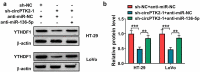
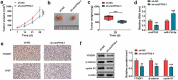
The dysregulated circular RNAs (circRNAs) are linked to progression and chemoresistance in colorectal cancer (CRC). However, the role of circRNA protein tyrosine kinase 2 (circPTK2) in CRC progression and chemoresistance is uncertain. The circPTK2, microRNA (miR)-136-5p, m6A 'reader' protein YTH domain family protein 1 (YTHDF1), β-catenin and cyclin D1 abundances were examined via quantitative reverse transcription PCR or Western blotting. The progression was investigated by cell counting kit-8 (CCK-8), colony formation, transwell and xenograft analysis. The resistance to 5-fluorouracil (5-FU) and oxaliplatin was analyzed via detecting cell viability and apoptosis using CCK-8 analysis and flow cytometry. The binding relationship was examined through dual-luciferase reporter, RNA immunoprecipitation and pull-down analysis. In our study, circPTK2 abundance was enhanced in CRC and associated with liver metastasis, clinical stage and chemoresistance. CircPTK2 knockdown constrained cell proliferation, migration, invasion, resistance to 5-FU and oxaliplatin, and the Wnt/β-catenin signaling. MiR-136-5p was bound with circPTK2 and downregulated in CRC. MiR-136-5p knockdown attenuated the influence of circPTK2 silence on CRC progression and chemoresistance. YTHDF1 was targeted via miR-136-5p and upregulated in CRC samples and cells. MiR-136-5p targeted YTHDF1 to restrain CRC progression and chemoresistance. In addition, we confirmed that circPTK2 silence reduced xenograft tumor growth. In conclusion, circPTK2 interference suppressed CRC proliferation, migration, invasion and chemoresistance via regulating miR-136-5p and YTHDF1.Abbreviations: circRNAs: circular RNAs; CRC: colorectal cancer; circPTK2: circRNA protein tyrosine kinase 2; miR: microRNA; YTHDF1: YTH domain family protein 1; CCK-8: cell counting kit-8; 5-FU: 5-fluorouracil; RIP: RNA immunoprecipitation.
Keywords: Colorectal cancer; YTHDF1; circPTK2; miR-136-5p.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
Similar articles
-
Circ_0000395 promotes cell growth, metastasis and oxaliplatin resistance by regulating miR-153-5p/MYO6 in colorectal cancer.Pathol Res Pract. 2024 Aug;260:155476. doi: 10.1016/j.prp.2024.155476. Epub 2024 Jul 17. Pathol Res Pract. 2024. PMID: 39038387
-
CircDDX17 reduces 5-fluorouracil resistance and hinders tumorigenesis in colorectal cancer by regulating miR-31-5p/KANK1 axis.Eur Rev Med Pharmacol Sci. 2020 Feb;24(4):1743-1754. doi: 10.26355/eurrev_202002_20351. Eur Rev Med Pharmacol Sci. 2020. PMID: 32141542
-
Exosomal circEPB41L2 serves as a sponge for miR-21-5p and miR-942-5p to suppress colorectal cancer progression by regulating the PTEN/AKT signalling pathway.Eur J Clin Invest. 2021 Sep;51(9):e13581. doi: 10.1111/eci.13581. Epub 2021 May 22. Eur J Clin Invest. 2021. PMID: 34022068
-
Molecular functions of microRNAs in colorectal cancer: recent roles in proliferation, angiogenesis, apoptosis, and chemoresistance.Naunyn Schmiedebergs Arch Pharmacol. 2024 Aug;397(8):5617-5630. doi: 10.1007/s00210-024-03076-w. Epub 2024 Apr 15. Naunyn Schmiedebergs Arch Pharmacol. 2024. PMID: 38619588 Review.
-
The significance of exosomal non-coding RNAs (ncRNAs) in the metastasis of colorectal cancer and development of therapy resistance.Gene. 2025 Feb 10;937:149141. doi: 10.1016/j.gene.2024.149141. Epub 2024 Dec 4. Gene. 2025. PMID: 39643147 Review.
Cited by
-
Circular RNA PGPEP1 induces colorectal cancer malignancy and immune escape.Cell Cycle. 2023 Jul-Aug;22(14-16):1743-1758. doi: 10.1080/15384101.2023.2225923. Epub 2023 Jul 9. Cell Cycle. 2023. PMID: 37424115 Free PMC article.
-
Cancer metastasis under the magnifying glass of epigenetics and epitranscriptomics.Cancer Metastasis Rev. 2023 Dec;42(4):1071-1112. doi: 10.1007/s10555-023-10120-3. Epub 2023 Jun 28. Cancer Metastasis Rev. 2023. PMID: 37369946 Free PMC article. Review.
-
Decoding the epitranscriptome: a new frontier for cancer therapy and drug resistance.Cell Commun Signal. 2024 Oct 21;22(1):513. doi: 10.1186/s12964-024-01854-w. Cell Commun Signal. 2024. PMID: 39434167 Free PMC article. Review.
-
Luteolin impacts deoxyribonucleic acid repair by modulating the mitogen-activated protein kinase pathway in colorectal cancer.Bioengineered. 2022 Apr;13(4):10998-11011. doi: 10.1080/21655979.2022.2066926. Bioengineered. 2022. PMID: 35473479 Free PMC article.
-
Overexpression of circ PTK2 suppresses the progression of nonalcoholic fatty liver disease via the miR-200c/SIK2/PI3K/Akt axis.Kaohsiung J Med Sci. 2022 Sep;38(9):869-878. doi: 10.1002/kjm2.12568. Epub 2022 Jul 6. Kaohsiung J Med Sci. 2022. PMID: 35791807 Free PMC article.
References
-
- Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. - PubMed
-
- Keum N, Giovannucci E.. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16(12):713–732. - PubMed
-
- Esmaeili M, Keshani M, Vakilian M, et al. Role of non-coding RNAs as novel biomarkers for detection of colorectal cancer progression through interaction with the cell signaling pathways. Gene. 2020;753:144796. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
